Gene Page: RARG
Summary ?
| GeneID | 5916 |
| Symbol | RARG |
| Synonyms | NR1B3|RARC |
| Description | retinoic acid receptor gamma |
| Reference | MIM:180190|HGNC:HGNC:9866|Ensembl:ENSG00000172819|HPRD:01573|Vega:OTTHUMG00000048077 |
| Gene type | protein-coding |
| Map location | 12q13 |
| Pascal p-value | 0.013 |
| TADA p-value | 0.001 |
| Fetal beta | -1.111 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RARG | chr12 | 53608241 | T | A | NM_000966 NM_001042728 NM_001243730 NM_001243731 NM_001243732 | p.209K>* p.198K>* p.137K>* p.88K>* p.187K>* | nonsense nonsense nonsense nonsense nonsense | Schizophrenia | DNM:Xu_2012 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27364319 | 12 | 53626837 | RARG | 9.92E-5 | -0.378 | 0.028 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16955618 | chr15 | 29937543 | RARG | 5916 | 0.14 | trans |
Section II. Transcriptome annotation
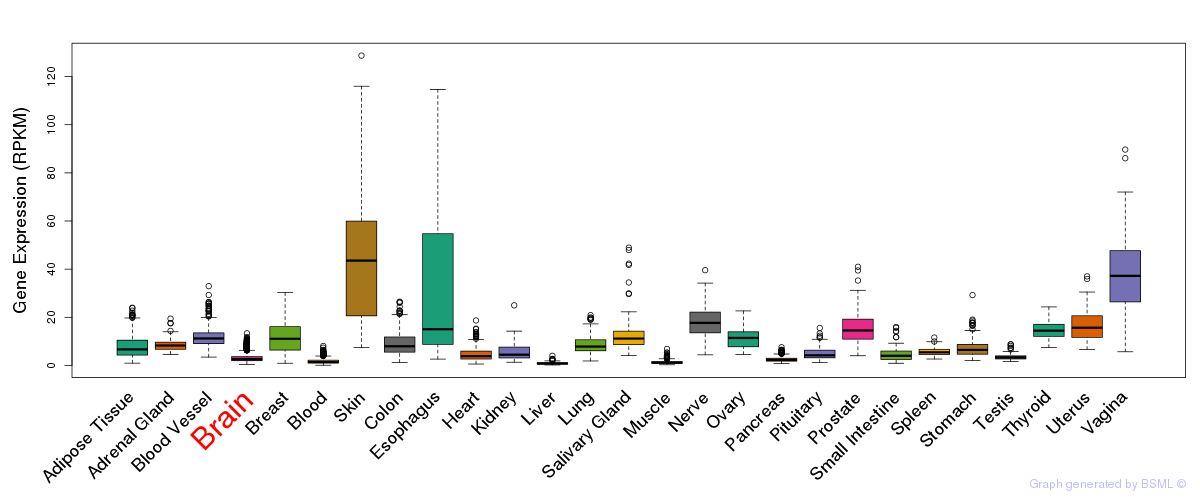
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDE6C | 0.50 | 0.29 |
| TAS2R4 | 0.50 | 0.41 |
| CILP | 0.50 | 0.44 |
| TAS2R48 | 0.50 | 0.43 |
| NAT8 | 0.49 | 0.39 |
| TGM1 | 0.48 | 0.41 |
| PLA2G4B | 0.48 | 0.39 |
| MST1R | 0.48 | 0.42 |
| AC034236.3 | 0.47 | 0.36 |
| SLC4A5 | 0.47 | 0.45 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IFI27 | -0.28 | -0.37 |
| RRAS | -0.28 | -0.37 |
| ITM2A | -0.28 | -0.35 |
| GNG11 | -0.27 | -0.38 |
| ABCG2 | -0.27 | -0.36 |
| RP11-884K10.1 | -0.27 | -0.36 |
| TM4SF18 | -0.27 | -0.33 |
| MT-CO2 | -0.26 | -0.34 |
| C1orf64 | -0.26 | -0.28 |
| S100A4 | -0.26 | -0.35 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0003708 | retinoic acid receptor activity | TAS | 2157970 | |
| GO:0005515 | protein binding | IPI | 10428834 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046965 | retinoid X receptor binding | ISS | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | ISS | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | ISS | - | |
| GO:0008361 | regulation of cell size | TAS | 18438858 | |
| GO:0048048 | embryonic eye morphogenesis | IEA | - | |
| GO:0007275 | multicellular organismal development | TAS | 2157970 | |
| GO:0043065 | positive regulation of apoptosis | IEA | - | |
| GO:0048384 | retinoic acid receptor signaling pathway | IDA | 17943189 | |
| GO:0032526 | response to retinoic acid | IDA | 17943189 | |
| GO:0035116 | embryonic hindlimb morphogenesis | ISS | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0060070 | Wnt receptor signaling pathway through beta-catenin | TAS | 18484682 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005667 | transcription factor complex | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 17943189 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| ZIRN TRETINOIN RESPONSE UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS UP | 92 | 64 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| SARTIPY BLUNTED BY INSULIN RESISTANCE DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MATTHEWS SKIN CARCINOGENESIS VIA JUN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION | 30 | 21 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| WANG NFKB TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 657 | 663 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-142-3p | 711 | 718 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-182 | 132 | 139 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA | ||||
| miR-24 | 410 | 416 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-30-5p | 1028 | 1035 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-326 | 822 | 828 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-331 | 884 | 890 | m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-34/449 | 273 | 279 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-96 | 133 | 139 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.