Gene Page: RARRES1
Summary ?
| GeneID | 5918 |
| Symbol | RARRES1 |
| Synonyms | LXNL|PERG-1|TIG1 |
| Description | retinoic acid receptor responder (tazarotene induced) 1 |
| Reference | MIM:605090|HGNC:HGNC:9867|Ensembl:ENSG00000118849|HPRD:05477|Vega:OTTHUMG00000158834 |
| Gene type | protein-coding |
| Map location | 3q25.32 |
| Pascal p-value | 0.973 |
| Fetal beta | 0.303 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08977270 | 3 | 158450833 | RARRES1 | 2.597E-4 | 0.322 | 0.038 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2274822 | chr6 | 14291088 | RARRES1 | 5918 | 0.19 | trans |
Section II. Transcriptome annotation
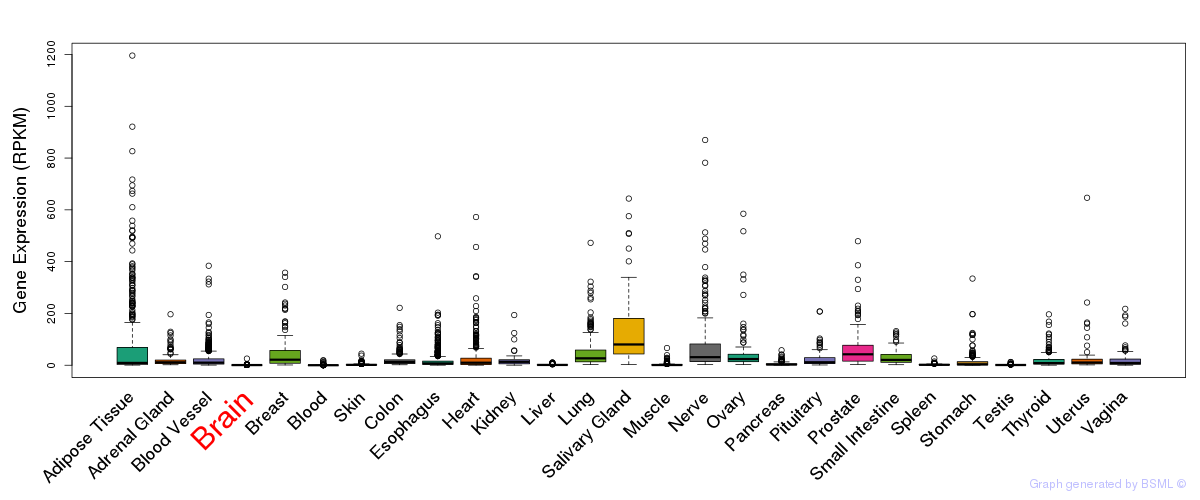
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ST6GALNAC6 | 0.90 | 0.91 |
| RUNDC3A | 0.85 | 0.84 |
| DDN | 0.84 | 0.87 |
| ADAP1 | 0.83 | 0.86 |
| FAM131A | 0.82 | 0.82 |
| NBL1 | 0.82 | 0.85 |
| ITPKA | 0.82 | 0.80 |
| RAB26 | 0.82 | 0.80 |
| LY6E | 0.82 | 0.79 |
| RHBDD2 | 0.81 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.55 | -0.50 |
| THOC2 | -0.55 | -0.59 |
| SH3BP2 | -0.54 | -0.61 |
| ATAD2B | -0.53 | -0.55 |
| UPF3B | -0.53 | -0.61 |
| RBMX2 | -0.53 | -0.61 |
| NASP | -0.53 | -0.55 |
| TUBB2B | -0.53 | -0.54 |
| IGF2BP3 | -0.53 | -0.49 |
| TIGD1 | -0.52 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTELIOMA SURVIVAL TIME DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL WORST VS BEST DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE UP | 52 | 36 | All SZGR 2.0 genes in this pathway |
| MOROSETTI FACIOSCAPULOHUMERAL MUSCULAR DISTROPHY UP | 20 | 11 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| ZIRN TRETINOIN RESPONSE WT1 UP | 23 | 14 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| APPEL IMATINIB RESPONSE | 33 | 22 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |