Gene Page: RHEB
Summary ?
| GeneID | 6009 |
| Symbol | RHEB |
| Synonyms | RHEB2 |
| Description | Ras homolog enriched in brain |
| Reference | MIM:601293|HGNC:HGNC:10011|Ensembl:ENSG00000106615|HPRD:03188|Vega:OTTHUMG00000157330 |
| Gene type | protein-coding |
| Map location | 7q36 |
| Pascal p-value | 0.023 |
| Fetal beta | -0.387 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14595269 | 7 | 151216272 | RHEB | 1.838E-4 | -0.227 | 0.034 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
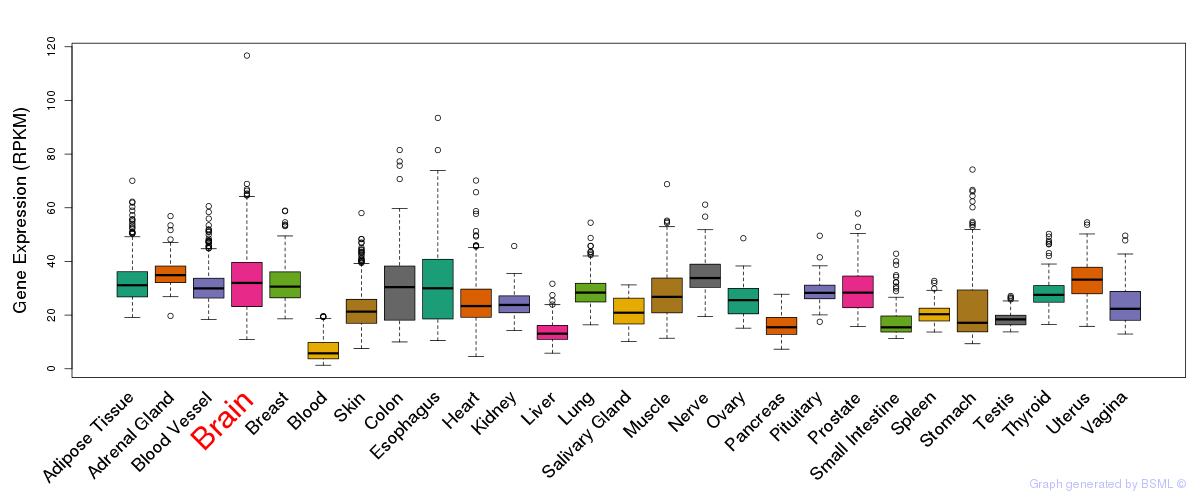
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA0494 | 0.83 | 0.84 |
| CLIP4 | 0.81 | 0.77 |
| NEBL | 0.81 | 0.79 |
| MAP3K5 | 0.81 | 0.81 |
| DLG1 | 0.80 | 0.79 |
| RNF6 | 0.80 | 0.76 |
| ARAP2 | 0.80 | 0.82 |
| CAST | 0.79 | 0.80 |
| ERMP1 | 0.79 | 0.83 |
| UTRN | 0.79 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.53 | -0.61 |
| RPL35 | -0.51 | -0.61 |
| RPL36 | -0.49 | -0.60 |
| RPL28 | -0.49 | -0.60 |
| SH2B2 | -0.48 | -0.46 |
| RPL27 | -0.48 | -0.57 |
| RPS19P3 | -0.47 | -0.61 |
| RPL18 | -0.47 | -0.60 |
| TRAF4 | -0.47 | -0.50 |
| RPLP1 | -0.46 | -0.56 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Affinity Capture-Western Reconstituted Complex | BioGRID | 15854902 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM interacts with Rheb. | BIND | 15854902 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | ATR interacts with Rheb. | BIND | 15854902 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Affinity Capture-Western Reconstituted Complex | BioGRID | 15854902 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Bcl-2 interacts with Rheb. | BIND | 9130713 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | FRAP1 (mTOR) interacts with Rheb. | BIND | 15854902 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15772076 |15854902 |15878852 |16915281 |
| GBL | LST8 | MGC111011 | POP3 | WAT1 | G protein beta subunit-like | GBL (LST8) interacts with Rheb. | BIND | 15854902 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Affinity Capture-Western | BioGRID | 15854902 |
| KIAA1303 | - | raptor | Raptor interacts with Rheb. | BIND | 15854902 |
| KIAA1303 | - | raptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 15854902 |
| PDE6D | PDED | phosphodiesterase 6D, cGMP-specific, rod, delta | - | HPRD,BioGRID | 11980706 |
| RAB7A | FLJ20819 | PRO2706 | RAB7 | RAB7A, member RAS oncogene family | Co-localization | BioGRID | 15809346 |
| RAB9A | RAB9 | RAB9A, member RAS oncogene family | Co-localization | BioGRID | 15809346 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 9001246 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | RAF interacts with Rheb. | BIND | 15854902 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Affinity Capture-Western | BioGRID | 15854902 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Biochemical Activity | BioGRID | 12820960 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD,BioGRID | 12842888 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | TSC2 interacts with Rheb. | BIND | 15854902 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MTORC1 MEDIATED SIGNALLING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP | 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| CORRADETTI MTOR PATHWAY REGULATORS DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |