Gene Page: RING1
Summary ?
| GeneID | 6015 |
| Symbol | RING1 |
| Synonyms | RING1A|RNF1 |
| Description | ring finger protein 1 |
| Reference | MIM:602045|HGNC:HGNC:10018|Ensembl:ENSG00000204227|HPRD:03624|Vega:OTTHUMG00000031278 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 6.658E-4 |
| Sherlock p-value | 0.004 |
| Fetal beta | -0 |
| DMG | 1 (# studies) |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12256228 | 6 | 33176613 | RING1 | -0.024 | 0.96 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
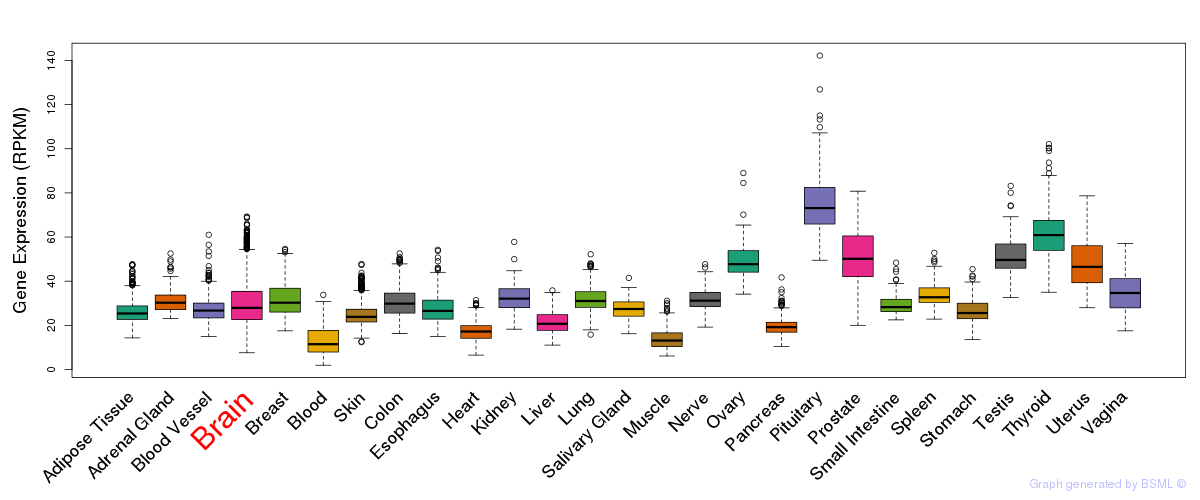
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRDM4 | 0.93 | 0.93 |
| DDX19A | 0.92 | 0.92 |
| EIF2AK4 | 0.92 | 0.91 |
| FAM120B | 0.92 | 0.92 |
| NOLC1 | 0.91 | 0.91 |
| ABCF2 | 0.91 | 0.90 |
| VPS39 | 0.91 | 0.93 |
| ZFYVE1 | 0.91 | 0.91 |
| RABGEF1 | 0.91 | 0.90 |
| RNF216 | 0.91 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.80 | -0.84 |
| MT-CO2 | -0.79 | -0.79 |
| AF347015.31 | -0.79 | -0.79 |
| AF347015.8 | -0.77 | -0.79 |
| FXYD1 | -0.77 | -0.78 |
| AF347015.33 | -0.75 | -0.75 |
| AF347015.27 | -0.75 | -0.77 |
| MT-CYB | -0.75 | -0.75 |
| HIGD1B | -0.75 | -0.76 |
| IFI27 | -0.73 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004842 | ubiquitin-protein ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | NAS | - | |
| GO:0016564 | transcription repressor activity | IDA | 9199346 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0016568 | chromatin modification | NAS | - | |
| GO:0016574 | histone ubiquitination | IEA | - | |
| GO:0048593 | camera-type eye morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000151 | ubiquitin ligase complex | IEA | - | |
| GO:0001739 | sex chromatin | IEA | - | |
| GO:0005634 | nucleus | IDA | 9199346 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0016607 | nuclear speck | IEA | - | |
| GO:0031519 | PcG protein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | RING1 interacts with BMI1. | BIND | 9858531 |
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | - | HPRD,BioGRID | 9199346 |9858531 |
| CBX2 | CDCA6 | M33 | MGC10561 | chromobox homolog 2 (Pc class homolog, Drosophila) | Reconstituted Complex | BioGRID | 10369680 |
| CBX4 | NBP16 | PC2 | hPC2 | chromobox homolog 4 (Pc class homolog, Drosophila) | RING1 interacts with HPC2. | BIND | 9858531 |
| CBX4 | NBP16 | PC2 | hPC2 | chromobox homolog 4 (Pc class homolog, Drosophila) | - | HPRD,BioGRID | 9199346 |9858531 |
| CBX7 | - | chromobox homolog 7 | - | HPRD,BioGRID | 14647293 |
| CBX8 | HPC3 | PC3 | RC1 | chromobox homolog 8 (Pc class homolog, Drosophila) | - | HPRD,BioGRID | 10825164 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Affinity Capture-Western | BioGRID | 11583618 |
| E2F6 | E2F-6 | MGC111545 | E2F transcription factor 6 | - | HPRD,BioGRID | 11171983 |
| ELAC2 | ELC2 | FLJ10530 | FLJ36693 | FLJ42848 | HPC2 | elaC homolog 2 (E. coli) | - | HPRD | 9199346 |
| FHL1 | FHL1B | FLH1A | KYO-T | MGC111107 | SLIM1 | XMPMA | bA535K18.1 | four and a half LIM domains 1 | KyoT interacts with RING1. This interaction was modeled on a demonstrated interaction between KyoT from unspecified species and human RING1. | BIND | 15710417 |
| PHC1 | EDR1 | HPH1 | RAE28 | polyhomeotic homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 9199346 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 11583618 |
| RING1 | RING1A | RNF1 | ring finger protein 1 | - | HPRD | 9199346 |
| RING1 | RING1A | RNF1 | ring finger protein 1 | Reconstituted Complex Two-hybrid | BioGRID | 9858531 |
| RING1 | RING1A | RNF1 | ring finger protein 1 | RING1 forms a homodimer. | BIND | 9858531 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | - | HPRD,BioGRID | 10369680 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STEGMEIER PRE-MITOTIC CELL CYCLE REGULATORS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 26 | 32 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.