Gene Page: RPA1
Summary ?
| GeneID | 6117 |
| Symbol | RPA1 |
| Synonyms | HSSB|MST075|REPA1|RF-A|RP-A|RPA70 |
| Description | replication protein A1 |
| Reference | MIM:179835|HGNC:HGNC:10289|Ensembl:ENSG00000132383|HPRD:01565|Vega:OTTHUMG00000090579 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 0.127 |
| Sherlock p-value | 0.761 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02316852 | 17 | 1737342 | RPA1 | 1.356E-4 | 0.259 | 0.031 | DMG:Wockner_2014 |
| cg22295628 | 17 | 1733433 | RPA1;SMYD4 | 1.932E-4 | -0.357 | 0.035 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
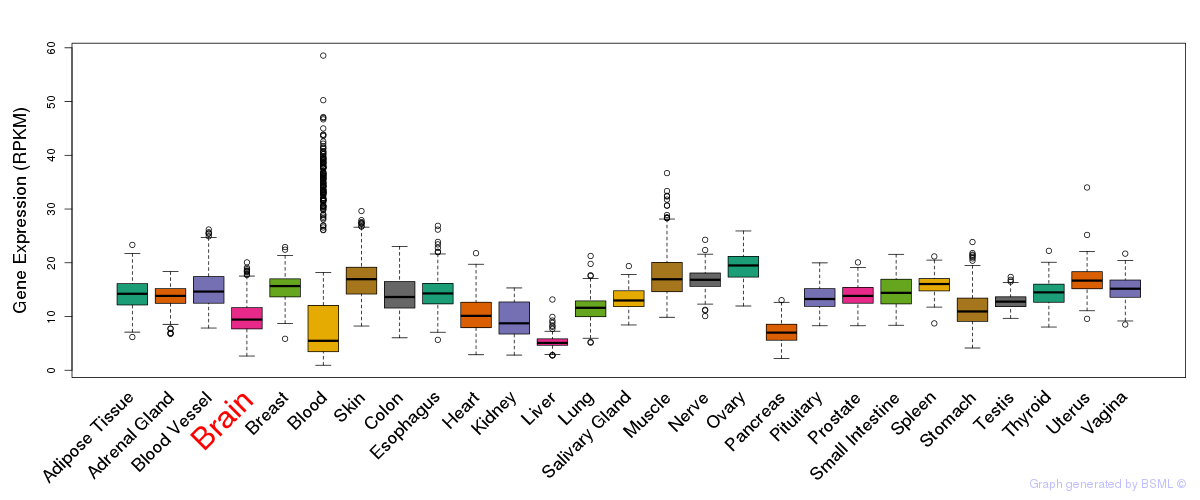
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKTIP | FT1 | FTS | AKT interacting protein | Two-hybrid | BioGRID | 16169070 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Protein-peptide | BioGRID | 10608806 |
| ATRIP | DKFZp762J2115 | FLJ12343 | MGC20625 | MGC21482 | MGC26740 | ATR interacting protein | Co-localization | BioGRID | 12791985 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | Biochemical Activity Co-localization Reconstituted Complex | BioGRID | 10825162 |11950880 |12181313 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | Affinity Capture-Western Reconstituted Complex | BioGRID | 12527904 |
| C8orf30A | FLJ40907 | chromosome 8 open reading frame 30A | Two-hybrid | BioGRID | 16169070 |
| CPE | - | carboxypeptidase E | Two-hybrid | BioGRID | 16169070 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Affinity Capture-MS | BioGRID | 12973351 |
| HELB | - | helicase (DNA) B | - | HPRD | 12181327 |
| KIN | BTCD | KIN17 | KIN, antigenic determinant of recA protein homolog (mouse) | Co-fractionation | BioGRID | 12754299 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD,BioGRID | 12614612 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | Two-hybrid | BioGRID | 12614612 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | Two-hybrid | BioGRID | 12614612 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD,BioGRID | 12614612 |
| MSH4 | - | mutS homolog 4 (E. coli) | Co-localization | BioGRID | 11950880 |
| MUTYH | MGC4416 | MYH | mutY homolog (E. coli) | hMYH is associated in vivo with replication protein A (RPA) | BIND | 11092888 |
| MUTYH | MGC4416 | MYH | mutY homolog (E. coli) | - | HPRD,BioGRID | 11092888 |
| ORC2L | ORC2 | origin recognition complex, subunit 2-like (yeast) | - | HPRD,BioGRID | 12614612 |
| ORC6L | ORC6 | origin recognition complex, subunit 6 like (yeast) | Two-hybrid | BioGRID | 12614612 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 12171929 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD,BioGRID | 10064605 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Reconstituted Complex | BioGRID | 12077133 |
| RAD52 | - | RAD52 homolog (S. cerevisiae) | Reconstituted Complex | BioGRID | 12077133 |
| RAD9A | RAD9 | RAD9 homolog A (S. pombe) | Rad9 interacts with RPA70. | BIND | 15897895 |
| RBM23 | CAPERbeta | FLJ10482 | MGC4458 | PP239 | RNPC4 | RNA binding motif protein 23 | Two-hybrid | BioGRID | 16169070 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | - | HPRD,BioGRID | 11927569 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | - | HPRD | 9461578 |12754299 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Affinity Capture-Western Co-crystal Structure | BioGRID | 10064605 |11927569 |
| RPA3 | REPA3 | replication protein A3, 14kDa | - | HPRD,BioGRID | 9461578 |
| RPA4 | HSU24186 | MGC120333 | MGC120334 | replication protein A4, 34kDa | - | HPRD,BioGRID | 7760808 |
| SELENBP1 | FLJ13813 | LPSB | SP56 | hSBP | hSP56 | selenium binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| TCEA2 | TFIIS | transcription elongation factor A (SII), 2 | Two-hybrid | BioGRID | 16169070 |
| TIPIN | FLJ20516 | TIMELESS interacting protein | Affinity Capture-Western Co-localization | BioGRID | 17296725 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11751427 |15489903 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15856006 |
| TREX1 | AGS1 | AGS5 | CRV | DKFZp434J0310 | DRN3 | HERNS | three prime repair exonuclease 1 | - | HPRD | 12791985 |
| XPA | XP1 | XPAC | xeroderma pigmentosum, complementation group A | - | HPRD,BioGRID | 7565690 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DNA REPLICATION | 36 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG NUCLEOTIDE EXCISION REPAIR | 44 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG MISMATCH REPAIR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| KEGG HOMOLOGOUS RECOMBINATION | 28 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION COUPLED NER TC NER | 45 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE EXCISION REPAIR | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME GLOBAL GENOMIC NER GG NER | 35 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF INCISION COMPLEX IN GG NER | 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME LAGGING STRAND SYNTHESIS | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | 38 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE | 75 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTENSION OF TELOMERES | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA STRAND ELONGATION | 30 | 18 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 12P11 12 UP | 33 | 17 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA TARGETS UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| OXFORD RALB TARGETS DN | 9 | 6 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA AND RALB TARGETS UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| FERRANDO HOX11 NEIGHBORS | 23 | 13 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| ISHIDA E2F TARGETS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER WITH BRCA1 MUTATED UP | 56 | 27 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |