Gene Page: RPL5
Summary ?
| GeneID | 6125 |
| Symbol | RPL5 |
| Synonyms | DBA6|L5|MSTP030|PPP1R135 |
| Description | ribosomal protein L5 |
| Reference | MIM:603634|HGNC:HGNC:10360|Ensembl:ENSG00000122406|HPRD:04699|Vega:OTTHUMG00000010899 |
| Gene type | protein-coding |
| Map location | 1p22.1 |
| Pascal p-value | 0.061 |
| Sherlock p-value | 0.179 |
| Fetal beta | 0.999 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0064 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RPL5 | chr1 | 93307363 | C | T | NM_000969 NM_001252273 | p.279R>W . | missense intronic | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
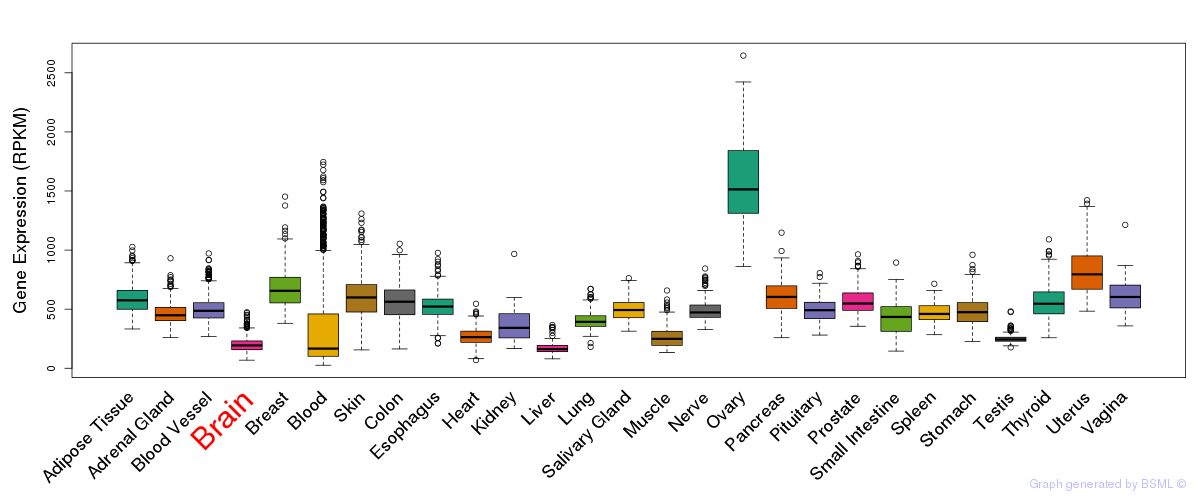
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPS25P8 | 0.96 | 0.96 |
| RPL38 | 0.96 | 0.94 |
| RPL37A | 0.96 | 0.95 |
| RPS24 | 0.95 | 0.94 |
| RPS8 | 0.95 | 0.95 |
| RPL23 | 0.94 | 0.94 |
| RPS27A | 0.94 | 0.90 |
| RPL26 | 0.94 | 0.91 |
| RPL30 | 0.94 | 0.94 |
| RPS23 | 0.94 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATP10A | -0.61 | -0.79 |
| BCL2L2 | -0.59 | -0.82 |
| SYNM | -0.57 | -0.79 |
| MAP3K5 | -0.57 | -0.75 |
| PTPRB | -0.57 | -0.74 |
| ACSL6 | -0.57 | -0.76 |
| EPB41L2 | -0.57 | -0.76 |
| LDHD | -0.56 | -0.71 |
| VWA5B2 | -0.56 | -0.78 |
| PK4P | -0.56 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003735 | structural constituent of ribosome | IEA | - | |
| GO:0003735 | structural constituent of ribosome | TAS | 7772601 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9465063 |12054647 | |
| GO:0008097 | 5S rRNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006414 | translational elongation | EXP | 15189156 | |
| GO:0006412 | translation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 14567916 | |
| GO:0005840 | ribosome | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0022625 | cytosolic large ribosomal subunit | TAS | 7772601 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 10491318 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD | 8806611 |9042965 |9738462 |10491318 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | Two-hybrid | BioGRID | 8806611 |9042965 |11710515 |14667819 |
| EIF5A | A | EIF-5A | EIF5A1 | MGC104255 | MGC99547 | uORF | eukaryotic translation initiation factor 5A | - | HPRD,BioGRID | 9465063 |
| HARS | FLJ20491 | HRS | histidyl-tRNA synthetase | - | HPRD,BioGRID | 8049265 |
| IPO13 | IMP13 | KAP13 | KIAA0724 | RANBP13 | importin 13 | Reconstituted Complex | BioGRID | 11447110 |
| IPO5 | DKFZp686O1576 | FLJ43041 | IMB3 | KPNB3 | MGC2068 | RANBP5 | importin 5 | - | HPRD,BioGRID | 9687515 |
| IPO7 | FLJ14581 | Imp7 | MGC138673 | RANBP7 | importin 7 | - | HPRD,BioGRID | 9687515 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 7935455 |14612427 |
| PDCD4 | H731 | MGC33046 | MGC33047 | programmed cell death 4 (neoplastic transformation inhibitor) | - | HPRD,BioGRID | 12054647 |
| RPL5 | MGC117339 | MSTP030 | ribosomal protein L5 | Protein-RNA Two-hybrid | BioGRID | 10491318 |15231747 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD,BioGRID | 9687515 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIBOSOME | 88 | 43 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | 179 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE CHAIN ELONGATION | 153 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A UP | 77 | 49 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| FLOTHO PEDIATRIC ALL THERAPY RESPONSE UP | 53 | 33 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS DN | 47 | 30 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM AND RAPAMYCIN SENSITIVE GENES | 68 | 35 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE B ALL UP | 26 | 15 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-376 | 13 | 19 | m8 | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.