Gene Page: RPL8
Summary ?
| GeneID | 6132 |
| Symbol | RPL8 |
| Synonyms | L8 |
| Description | ribosomal protein L8 |
| Reference | MIM:604177|HGNC:HGNC:10368|Ensembl:ENSG00000161016|HPRD:16044|Vega:OTTHUMG00000165249 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.078 |
| Sherlock p-value | 0.168 |
| Fetal beta | 0.91 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26911140 | 8 | 146017711 | RPL8 | -0.019 | 0.65 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
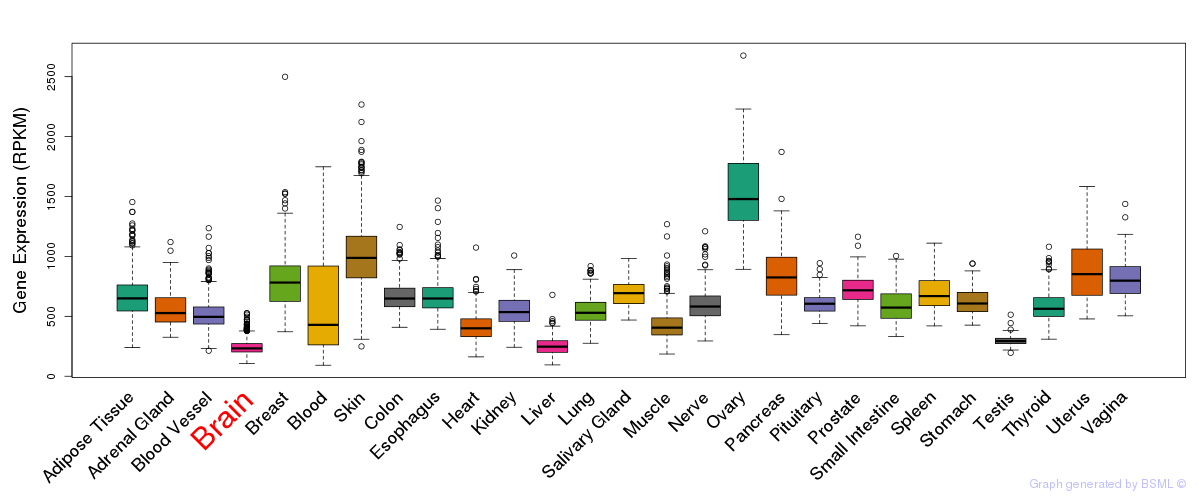
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPL26 | 0.95 | 0.94 |
| RPS20 | 0.94 | 0.93 |
| RPS27A | 0.93 | 0.92 |
| RPL38 | 0.93 | 0.89 |
| RPL27A | 0.92 | 0.88 |
| RPL30 | 0.92 | 0.92 |
| RPL35A | 0.92 | 0.92 |
| RPS24 | 0.92 | 0.91 |
| RPL32 | 0.92 | 0.93 |
| RPL27 | 0.92 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNM | -0.57 | -0.77 |
| PTPRB | -0.57 | -0.74 |
| ATP10A | -0.57 | -0.73 |
| EPB41L2 | -0.56 | -0.74 |
| BCL2L2 | -0.56 | -0.74 |
| EHD2 | -0.55 | -0.76 |
| SEMA3G | -0.55 | -0.71 |
| SLC20A2 | -0.55 | -0.71 |
| EDN3 | -0.55 | -0.69 |
| PK4P | -0.55 | -0.65 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIBOSOME | 88 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | 179 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE CHAIN ELONGATION | 153 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| AUNG GASTRIC CANCER | 54 | 20 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM AND RAPAMYCIN SENSITIVE GENES | 68 | 35 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |