Gene Page: RXRA
Summary ?
| GeneID | 6256 |
| Symbol | RXRA |
| Synonyms | NR2B1 |
| Description | retinoid X receptor alpha |
| Reference | MIM:180245|HGNC:HGNC:10477|HPRD:01577| |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.498 |
| Sherlock p-value | 0.57 |
| Fetal beta | -1.106 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
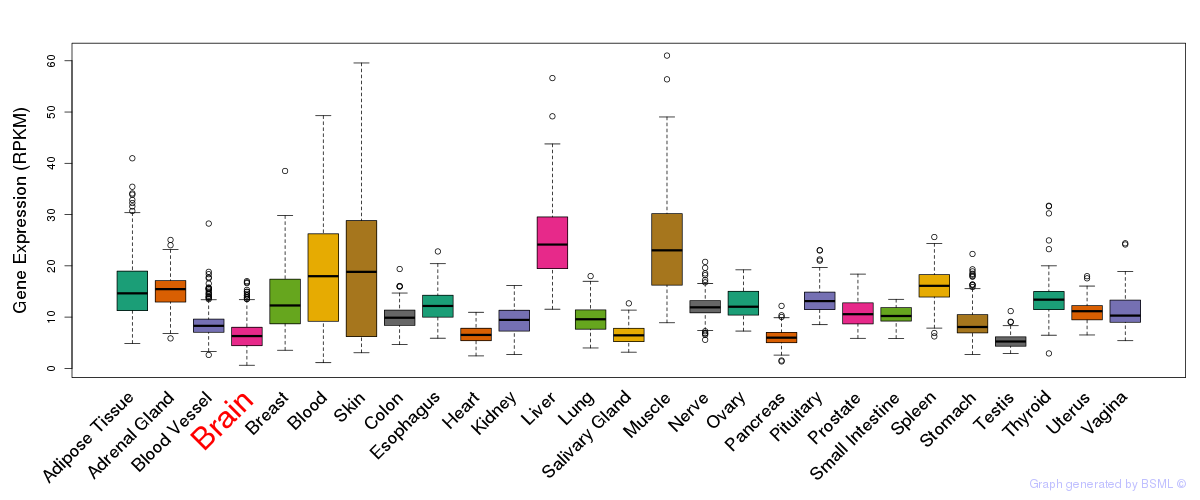
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004886 | retinoid-X receptor activity | TAS | 1310260 | |
| GO:0003674 | molecular_function | ND | - | |
| GO:0003700 | transcription factor activity | IDA | 12037571 | |
| GO:0003706 | ligand-regulated transcription factor activity | IDA | 7990953 | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0003713 | transcription coactivator activity | TAS | 1651173 | |
| GO:0005496 | steroid binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 12040021 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055012 | ventricular cardiac muscle cell differentiation | IEA | - | |
| GO:0055010 | ventricular cardiac muscle morphogenesis | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008203 | cholesterol metabolic process | TAS | 18544046 | |
| GO:0008150 | biological_process | ND | - | |
| GO:0006766 | vitamin metabolic process | TAS | 1651173 | |
| GO:0048384 | retinoic acid receptor signaling pathway | IMP | 17538076 | |
| GO:0032526 | response to retinoic acid | IMP | 17538076 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 12037571 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0060038 | cardiac muscle cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005634 | nucleus | IDA | 7990953 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | Reconstituted Complex | BioGRID | 11439184 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 9812988 |
| BRD8 | SMAP | SMAP2 | p120 | bromodomain containing 8 | Reconstituted Complex Two-hybrid | BioGRID | 10517671 |
| CLOCK | KAT13D | KIAA0334 | bHLHe8 | clock homolog (mouse) | Reconstituted Complex Two-hybrid | BioGRID | 11439184 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD | 7776974 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 12771132 |
| CTSL1 | CATL | CTSL | FLJ31037 | MEP | cathepsin L1 | - | HPRD,BioGRID | 9837884 |9918848 |
| DNTTIP2 | ERBP | FCF2 | HSU15552 | LPTS-RP2 | MGC163494 | RP4-561L24.1 | TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 | - | HPRD,BioGRID | 15047147 |
| EDF1 | EDF-1 | MBF1 | MGC9058 | endothelial differentiation-related factor 1 | - | HPRD,BioGRID | 12040021 |
| FUS | CHOP | FUS-CHOP | FUS1 | TLS | TLS/CHOP | hnRNP-P2 | fusion (involved in t(12;16) in malignant liposarcoma) | - | HPRD,BioGRID | 9440806 |
| GADD45A | DDIT1 | GADD45 | growth arrest and DNA-damage-inducible, alpha | - | HPRD,BioGRID | 10872826 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD,BioGRID | 10872826 |
| HBXIP | MGC71071 | XIP | hepatitis B virus x interacting protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 14578865 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Reconstituted Complex | BioGRID | 12943985 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Reconstituted Complex | BioGRID | 12943985 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 10874028 |
| ITGB3BP | CENP-R | CENPR | HSU37139 | NRIF3 | TAP20 | integrin beta 3 binding protein (beta3-endonexin) | - | HPRD,BioGRID | 10490654 |
| JMJD1C | DKFZp761F0118 | FLJ14374 | KIAA1380 | RP11-10C13.2 | TRIP8 | jumonji domain containing 1C | - | HPRD | 7776974 |
| MECR | CGI-63 | FASN2B | NRBF1 | mitochondrial trans-2-enoyl-CoA reductase | - | HPRD | 9795230 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 9653119 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | - | HPRD,BioGRID | 9653119 |
| MPG | AAG | APNG | CRA36.1 | MDG | Mid1 | PIG11 | PIG16 | anpg | N-methylpurine-DNA glycosylase | MPG interacts with RXRA (RXR-alpha). This interaction was modeled on a demonstrated interaction between MPG from an unspecified species and RXRA from an unspecified species. | BIND | 14761960 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 9692544 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | Two-hybrid | BioGRID | 12612084 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 7481822 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | GRIP1 interacts with RXR. This interaction was modeled on a demonstrated interaction between mouse GRIP1 and human RXR. | BIND | 9920895 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | Reconstituted Complex Two-hybrid | BioGRID | 11851396 |12612084 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | ACTR interacts with RXR. | BIND | 9267036 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD,BioGRID | 9267036 |
| NCOA4 | ARA70 | DKFZp762E1112 | ELE1 | PTC3 | RFG | nuclear receptor coactivator 4 | - | HPRD | 10347167 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10567404 |11158331 |11773444 |14578865 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 10075655 |
| NFKBIB | IKBB | TRIP9 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | - | HPRD,BioGRID | 7776974 |9452433 |
| NPAS2 | FLJ23138 | MGC71151 | MOP4 | PASD4 | bHLHe9 | neuronal PAS domain protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 11439184 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | - | HPRD,BioGRID | 10594021 |
| NR1H2 | LXR-b | LXRB | NER | NER-I | RIP15 | UNR | nuclear receptor subfamily 1, group H, member 2 | - | HPRD,BioGRID | 7760852 |
| NR1H3 | LXR-a | LXRA | RLD-1 | nuclear receptor subfamily 1, group H, member 3 | Reconstituted Complex | BioGRID | 8621574 |
| NR1H4 | BAR | FXR | HRR-1 | HRR1 | MGC163445 | RIP14 | nuclear receptor subfamily 1, group H, member 4 | - | HPRD,BioGRID | 7760852 |
| NR1I2 | BXR | ONR1 | PAR | PAR1 | PAR2 | PARq | PRR | PXR | SAR | SXR | nuclear receptor subfamily 1, group I, member 2 | - | HPRD | 9727070 |
| NR1I3 | CAR | CAR1 | MB67 | MGC150433 | MGC97144 | MGC97209 | nuclear receptor subfamily 1, group I, member 3 | - | HPRD,BioGRID | 8114692 |
| NR2E3 | ESCS | MGC49976 | PNR | RNR | RP37 | rd7 | nuclear receptor subfamily 2, group E, member 3 | Reconstituted Complex | BioGRID | 10611353 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD | 1311101 |
| NR2F6 | EAR-2 | EAR2 | ERBAL2 | nuclear receptor subfamily 2, group F, member 6 | Two-hybrid | BioGRID | 10318855 |
| NR4A1 | GFRP1 | HMR | MGC9485 | N10 | NAK-1 | NGFIB | NP10 | NUR77 | TR3 | nuclear receptor subfamily 4, group A, member 1 | Reconstituted Complex Two-hybrid | BioGRID | 14980220 |
| NR4A2 | HZF-3 | NOT | NURR1 | RNR1 | TINUR | nuclear receptor subfamily 4, group A, member 2 | - | HPRD | 7705655 |7758108 |
| NRBF2 | COPR1 | COPR2 | DKFZp564C1664 | FLJ30395 | NRBF-2 | nuclear receptor binding factor 2 | - | HPRD,BioGRID | 10786636 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD,BioGRID | 8887632 |
| NSD1 | ARA267 | DKFZp666C163 | FLJ10684 | FLJ22263 | FLJ44628 | KMT3B | SOTOS | STO | nuclear receptor binding SET domain protein 1 | - | HPRD | 9628876 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD | 10082530 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Reconstituted Complex | BioGRID | 10610177 |
| PNRC2 | FLJ20312 | MGC99541 | proline-rich nuclear receptor coactivator 2 | - | HPRD | 11574675 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD,BioGRID | 10383413 |
| POU2F2 | OCT2 | OTF2 | Oct-2 | POU class 2 homeobox 2 | Reconstituted Complex | BioGRID | 10383413 |
| PPARA | MGC2237 | MGC2452 | NR1C1 | PPAR | hPPAR | peroxisome proliferator-activated receptor alpha | - | HPRD,BioGRID | 10195690 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD,BioGRID | 7838715 |10854698 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD | 7838715 |10854698 |10882139 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | - | HPRD,BioGRID | 11714715 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with RXR-alpha. | BIND | 12039952 |
| PSMC3IP | GT198 | HOP2 | HUMGT198A | TBPIP | PSMC3 interacting protein | - | HPRD | 11739747 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Co-purification Reconstituted Complex | BioGRID | 1314167 |12052862 |
| RARG | NR1B3 | RARC | retinoic acid receptor, gamma | - | HPRD | 10835357 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Reconstituted Complex | BioGRID | 10075655 |
| RNF8 | FLJ12013 | KIAA0646 | ring finger protein 8 | - | HPRD,BioGRID | 14981089 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 10361124 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10517671 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD | 11641790 |
| TADA3L | ADA3 | FLJ20221 | FLJ21329 | hADA3 | transcriptional adaptor 3 (NGG1 homolog, yeast)-like | - | HPRD,BioGRID | 12235159 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 7667283 |
| TDG | - | thymine-DNA glycosylase | Reconstituted Complex | BioGRID | 12874288 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Co-purification | BioGRID | 1314167 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | - | HPRD | 1331778 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | Reconstituted Complex | BioGRID | 9368056 |
| TMPRSS3 | DFNB10 | DFNB8 | ECHOS1 | TADG12 | transmembrane protease, serine 3 | Two-hybrid | BioGRID | 16169070 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD | 9115274 |9632676 |11851396 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | Reconstituted Complex | BioGRID | 9115274 |11851396 |
| TRIP10 | CIP4 | HSTP | STOT | STP | thyroid hormone receptor interactor 10 | - | HPRD | 7776974 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD | 7776974 |
| UBQLN4 | A1U | C1orf6 | CIP75 | UBIN | ubiquilin 4 | Two-hybrid | BioGRID | 16169070 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Reconstituted Complex Two-hybrid | BioGRID | 9632709 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | VDR interacts with RXRA (RXRalpha) to form a heterodimer. | BIND | 15829977 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD | 8622986 |
| ZNHIT3 | TRIP3 | zinc finger, HIT type 3 | - | HPRD | 7776974 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VDR PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RARRXR PATHWAY | 15 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM1 PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| EINAV INTERFERON SIGNATURE IN CANCER | 27 | 16 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS UP | 13 | 9 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| GOTTWEIN TARGETS OF KSHV MIR K12 11 | 63 | 45 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN | 56 | 39 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY DN | 10 | 10 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 3381 | 3387 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 2093 | 2099 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 3804 | 3810 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 1431 | 1437 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-148/152 | 1432 | 1438 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-19 | 1430 | 1436 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-27 | 3804 | 3811 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-493-5p | 3498 | 3504 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-9 | 623 | 629 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.