Gene Page: CDH23
Summary ?
| GeneID | 64072 |
| Symbol | CDH23 |
| Synonyms | CDHR23|USH1D |
| Description | cadherin-related 23 |
| Reference | MIM:605516|HGNC:HGNC:13733|Ensembl:ENSG00000107736|HPRD:05699|Vega:OTTHUMG00000019347 |
| Gene type | protein-coding |
| Map location | 10q22.1 |
| Pascal p-value | 0.244 |
| Sherlock p-value | 0.789 |
| Fetal beta | -1.583 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12568633 | 10 | 73533407 | CDH23;C10orf54 | 3.36E-5 | -0.305 | 0.019 | DMG:Wockner_2014 |
| cg09810750 | 10 | 73533449 | CDH23 | 4.41E-8 | -0.016 | 1.21E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | CDH23 | 64072 | 3.913E-5 | trans | ||
| rs6741728 | chr2 | 153268745 | CDH23 | 64072 | 0.19 | trans | ||
| rs7584986 | chr2 | 184111432 | CDH23 | 64072 | 0.02 | trans | ||
| rs1396222 | chr4 | 173279496 | CDH23 | 64072 | 0.16 | trans | ||
| rs335993 | chr4 | 173321789 | CDH23 | 64072 | 0.18 | trans | ||
| rs10491487 | chr5 | 80323367 | CDH23 | 64072 | 3.997E-4 | trans | ||
| rs17057688 | chr6 | 130033232 | CDH23 | 64072 | 0 | trans | ||
| rs6996695 | chr8 | 77540580 | CDH23 | 64072 | 0.02 | trans | ||
| rs11139334 | chr9 | 84209393 | CDH23 | 64072 | 3.532E-5 | trans | ||
| snp_a-4267673 | 0 | CDH23 | 64072 | 0.01 | trans | |||
| rs3762058 | chr9 | 116842332 | CDH23 | 64072 | 0.17 | trans | ||
| rs946861 | chr10 | 21281250 | CDH23 | 64072 | 0.19 | trans | ||
| rs7111644 | chr11 | 93300259 | CDH23 | 64072 | 0.15 | trans | ||
| rs9989228 | chr14 | 37809252 | CDH23 | 64072 | 0.12 | trans | ||
| rs16955618 | chr15 | 29937543 | CDH23 | 64072 | 1.092E-5 | trans | ||
| rs2077735 | chr15 | 58479024 | CDH23 | 64072 | 0.04 | trans |
Section II. Transcriptome annotation
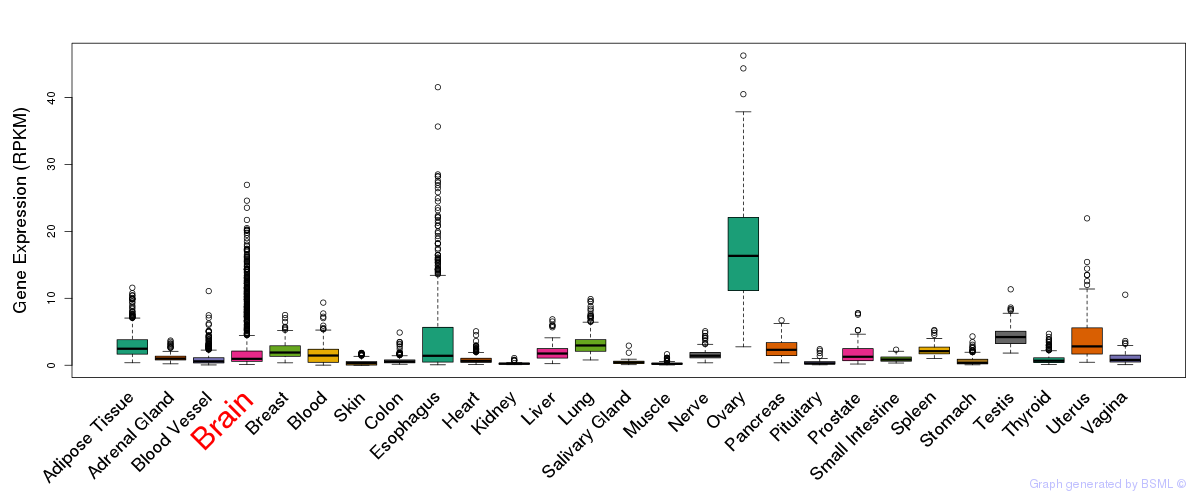
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |