Gene Page: SRSF1
Summary ?
| GeneID | 6426 |
| Symbol | SRSF1 |
| Synonyms | ASF|SF2|SF2p33|SFRS1|SRp30a |
| Description | serine/arginine-rich splicing factor 1 |
| Reference | MIM:600812|HGNC:HGNC:10780|HPRD:02887| |
| Gene type | protein-coding |
| Map location | 17q22 |
| Pascal p-value | 0.568 |
| Sherlock p-value | 0.187 |
| Fetal beta | 1.007 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24063238 | 17 | 56085336 | SRSF1 | 1.79E-8 | -0.021 | 6.42E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
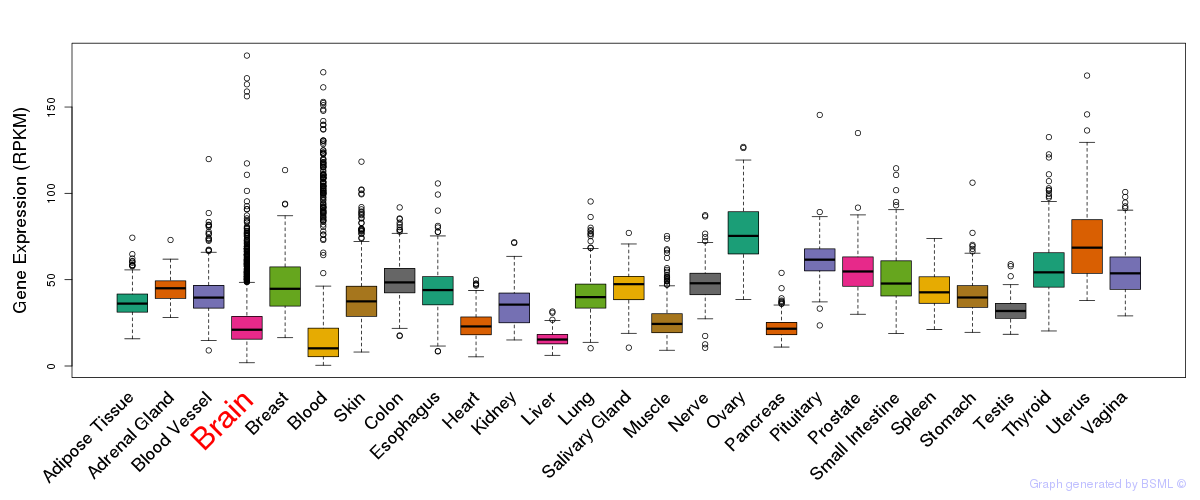
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM54 | 0.66 | 0.71 |
| C5orf32 | 0.66 | 0.66 |
| LRRC29 | 0.65 | 0.65 |
| GCAT | 0.64 | 0.64 |
| CIB1 | 0.64 | 0.67 |
| CISD3 | 0.64 | 0.64 |
| ST6GALNAC6 | 0.62 | 0.62 |
| RAMP1 | 0.62 | 0.65 |
| ISOC2 | 0.62 | 0.59 |
| C17orf89 | 0.61 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAP4K4 | -0.51 | -0.56 |
| THOC2 | -0.50 | -0.55 |
| CTTNBP2NL | -0.50 | -0.53 |
| ZCCHC6 | -0.49 | -0.54 |
| ZNF197 | -0.49 | -0.51 |
| PHF16 | -0.49 | -0.54 |
| ZNF776 | -0.49 | -0.51 |
| PHF20 | -0.49 | -0.52 |
| LRIG2 | -0.49 | -0.52 |
| BAZ1B | -0.49 | -0.53 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C1QBP | GC1QBP | HABP1 | SF2p32 | gC1Q-R | gC1qR | p32 | complement component 1, q subcomponent binding protein | - | HPRD,BioGRID | 10022843 |
| CLK1 | CLK | CLK/STY | STY | CDC-like kinase 1 | Biochemical Activity | BioGRID | 8798720 |12565863 |
| CLK1 | CLK | CLK/STY | STY | CDC-like kinase 1 | - | HPRD | 8617202 |
| CRKRS | CRK7 | CRKR | KIAA0904 | Cdc2-related kinase, arginine/serine-rich | - | HPRD | 11683387 |
| CROP | LUC7A | OA48-18 | cisplatin resistance-associated overexpressed protein | - | HPRD,BioGRID | 12565863 |
| DIDO1 | BYE1 | C20orf158 | DATF1 | DIDO2 | DIDO3 | DIO-1 | DIO1 | DKFZp434P1115 | FLJ11265 | KIAA0333 | MGC16140 | dJ885L7.8 | death inducer-obliterator 1 | Two-hybrid | BioGRID | 16189514 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | Biochemical Activity | BioGRID | 14623875 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| PRPF4 | HPRP4 | HPRP4P | PRP4 | Prp4p | PRP4 pre-mRNA processing factor 4 homolog (yeast) | - | HPRD,BioGRID | 11418604 |
| PRPF4B | KIAA0536 | PR4H | PRP4 | PRP4H | PRP4K | dJ1013A10.1 | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | - | HPRD | 11418604 |
| PSIP1 | DFS70 | LEDGF | MGC74712 | PAIP | PSIP2 | p52 | p75 | PC4 and SFRS1 interacting protein 1 | - | HPRD,BioGRID | 9885563 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Reconstituted Complex Two-hybrid | BioGRID | 8816452 |9799243 |
| SFRS11 | DKFZp686M13204 | dJ677H15.2 | p54 | splicing factor, arginine/serine-rich 11 | - | HPRD,BioGRID | 8816452 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 10757789 |14559993 |
| SFRS2 | PR264 | SC-35 | SC35 | SFRS2A | SRp30b | splicing factor, arginine/serine-rich 2 | Two-hybrid | BioGRID | 8816452 |
| SFRS2IP | CASP11 | SIP1 | SRRP129 | splicing factor, arginine/serine-rich 2, interacting protein | - | HPRD,BioGRID | 9447963 |
| SFRS5 | HRS | SRP40 | splicing factor, arginine/serine-rich 5 | Reconstituted Complex | BioGRID | 9799243 |
| SIP1 | GEMIN2 | SIP1-delta | survival of motor neuron protein interacting protein 1 | - | HPRD | 9447963 |
| SNRNP70 | RNPU1Z | RPU1 | SNRP70 | U170K | U1AP | U1RNP | small nuclear ribonucleoprotein 70kDa (U1) | - | HPRD,BioGRID | 9799243 |
| SRPK1 | SFRSK1 | SFRS protein kinase 1 | - | HPRD | 8208298 |8798720 |9472028 |10390541 |
| SRPK1 | SFRSK1 | SFRS protein kinase 1 | Affinity Capture-Western Biochemical Activity Far Western Reconstituted Complex | BioGRID | 9472028 |10196197 |12565863 |17517895 |
| SRPK2 | FLJ36101 | SFRSK2 | SFRS protein kinase 2 | - | HPRD,BioGRID | 9472028 |10196197 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
| TNPO3 | IPO12 | MTR10A | TRN-SR | TRN-SR2 | TRNSR | transportin 3 | - | HPRD | 10366588 |10713112 |
| TOP1 | TOPI | topoisomerase (DNA) I | - | HPRD | 9611241|12270705 |
| TOP1 | TOPI | topoisomerase (DNA) I | Far Western Reconstituted Complex | BioGRID | 9611241 |12270705 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | - | HPRD | 8261509 |9799243 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | Reconstituted Complex Two-hybrid | BioGRID | 8816452 |9799243 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | Two-hybrid | BioGRID | 16189514 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | - | HPRD | 9473574 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| ZRSR2 | MGC142014 | MGC142040 | U2AF1-RS2 | U2AF1L2 | U2AF1RS2 | URP | zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 | - | HPRD | 9237760 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA 3 END PROCESSING | 35 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG DN | 59 | 40 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A UP | 77 | 49 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY DN | 15 | 8 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS DN | 60 | 45 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8 | 49 | 36 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |