Gene Page: ARAP3
Summary ?
| GeneID | 64411 |
| Symbol | ARAP3 |
| Synonyms | CENTD3|DRAG1 |
| Description | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| Reference | MIM:606647|HGNC:HGNC:24097|Ensembl:ENSG00000120318|HPRD:09441|Vega:OTTHUMG00000129610 |
| Gene type | protein-coding |
| Map location | 5q31.3 |
| Pascal p-value | 0.687 |
| Sherlock p-value | 0.689 |
| Fetal beta | 0.228 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15043642 | 5 | 141061863 | ARAP3 | 6.07E-5 | -0.437 | 0.023 | DMG:Wockner_2014 |
| cg04477203 | 5 | 141061883 | ARAP3 | 1.285E-4 | -0.53 | 0.03 | DMG:Wockner_2014 |
| cg11893951 | 5 | 141061521 | ARAP3 | 3.271E-4 | -0.575 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12564436 | chr1 | 55057662 | ARAP3 | 64411 | 0.02 | trans | ||
| rs2289015 | chr1 | 55075061 | ARAP3 | 64411 | 0.1 | trans | ||
| rs892932 | chr3 | 11624210 | ARAP3 | 64411 | 0.19 | trans | ||
| rs6793958 | chr3 | 17853055 | ARAP3 | 64411 | 0.01 | trans | ||
| rs7635430 | chr3 | 180152035 | ARAP3 | 64411 | 0.02 | trans | ||
| rs16933426 | chr10 | 78113005 | ARAP3 | 64411 | 0.01 | trans | ||
| rs17115591 | chr14 | 45262026 | ARAP3 | 64411 | 0.15 | trans | ||
| rs689654 | chr15 | 54029474 | ARAP3 | 64411 | 0.11 | trans | ||
| rs2279706 | chr19 | 31799790 | ARAP3 | 64411 | 0.14 | trans | ||
| rs4812218 | chr20 | 59422845 | ARAP3 | 64411 | 0.12 | trans | ||
| rs17201877 | chr20 | 59435605 | ARAP3 | 64411 | 0.03 | trans | ||
| rs2822799 | chr21 | 15958167 | ARAP3 | 64411 | 0.14 | trans |
Section II. Transcriptome annotation
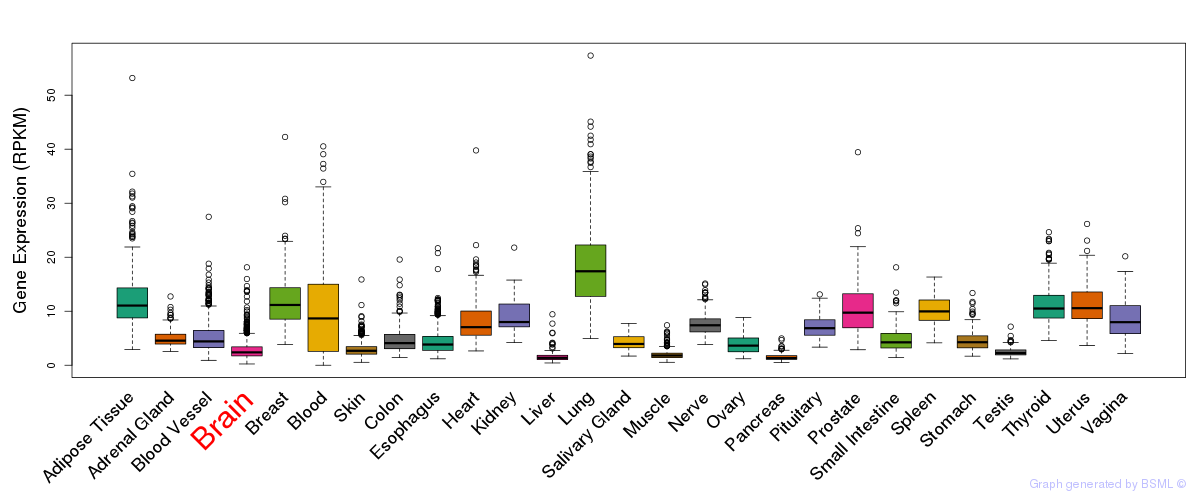
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005547 | phosphatidylinositol-3,4,5-triphosphate binding | IDA | 11804589 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008060 | ARF GTPase activator activity | ISS | - | |
| GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding | IDA | 11804589 | |
| GO:0030675 | Rac GTPase activator activity | ISS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007010 | cytoskeleton organization | TAS | 15569923 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008360 | regulation of cell shape | ISS | - | |
| GO:0016192 | vesicle-mediated transport | TAS | 15569923 | |
| GO:0035024 | negative regulation of Rho protein signal transduction | ISS | - | |
| GO:0032312 | regulation of ARF GTPase activity | IEA | - | |
| GO:0030336 | negative regulation of cell migration | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | ISS | - | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IC | 11804589 | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | ISS | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE UP | 26 | 19 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS DN | 53 | 29 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-150 | 482 | 488 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.