Gene Page: SH3GL3
Summary ?
| GeneID | 6457 |
| Symbol | SH3GL3 |
| Synonyms | CNSA3|EEN-B2|HsT19371|SH3D2C|SH3P13 |
| Description | SH3-domain GRB2-like 3 |
| Reference | MIM:603362|HGNC:HGNC:10832|Ensembl:ENSG00000140600|HPRD:04528|Vega:OTTHUMG00000147361 |
| Gene type | protein-coding |
| Map location | 15q24 |
| Pascal p-value | 2.666E-4 |
| Sherlock p-value | 0.95 |
| Fetal beta | -0.225 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0594 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2562784 | 15 | 84286492 | null | 1.674 | SH3GL3 | null |
Section II. Transcriptome annotation
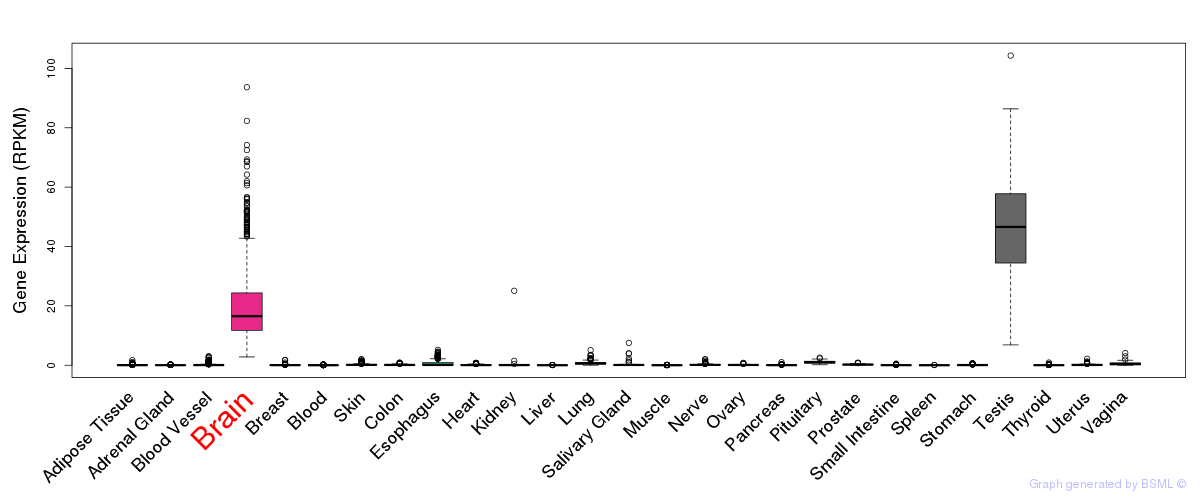
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0008289 | lipid binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 16115810 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 9169142 |
| GO:0007165 | signal transduction | TAS | 9169142 | |
| GO:0006897 | endocytosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005768 | endosome | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0031901 | early endosome membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | Beta-1-AR interacts with SH3p13. This interaction was modeled on a demonstrated interaction between human Beta-1-AR and rat SH3p13. | BIND | 10535961 |
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Two-hybrid | BioGRID | 16169070 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Two-hybrid | BioGRID | 16169070 |
| C11orf68 | BLES03 | P5326 | chromosome 11 open reading frame 68 | Two-hybrid | BioGRID | 16169070 |
| C4orf17 | DKFZp434G072 | chromosome 4 open reading frame 17 | Two-hybrid | BioGRID | 16169070 |
| CALCOCO1 | Cocoa | KIAA1536 | PP13275 | calphoglin | calcium binding and coiled-coil domain 1 | Two-hybrid | BioGRID | 16169070 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 11894096 |
| CRIP1 | CRHP | CRIP | CRP1 | FLJ40971 | cysteine-rich protein 1 (intestinal) | Two-hybrid | BioGRID | 16169070 |
| DNM1 | DNM | dynamin 1 | Endophilin A3 interacts with Dynamin. | BIND | 11894096 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 9238017 |
| DPPA4 | 2410091M23Rik | FLJ10713 | developmental pluripotency associated 4 | Two-hybrid | BioGRID | 16169070 |
| DPYSL4 | CRMP3 | DRP-4 | ULIP4 | dihydropyrimidinase-like 4 | Two-hybrid | BioGRID | 16169070 |
| EGFL6 | DKFZp564P2063 | MAEG | W80 | EGF-like-domain, multiple 6 | Two-hybrid | BioGRID | 16169070 |
| EMD | EDMD | LEMD5 | STA | emerin | Two-hybrid | BioGRID | 16169070 |
| FAM173A | C16orf24 | MGC2494 | family with sequence similarity 173, member A | Two-hybrid | BioGRID | 16169070 |
| GABBR2 | FLJ36928 | GABABR2 | GPR51 | GPRC3B | HG20 | HRIHFB2099 | gamma-aminobutyric acid (GABA) B receptor, 2 | Two-hybrid | BioGRID | 16169070 |
| HTT | HD | IT15 | huntingtin | HD interacts with SH3GL3. | BIND | 9809064 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 9809064 |
| IGSF2 | CD101 | V7 | immunoglobulin superfamily, member 2 | Two-hybrid | BioGRID | 16169070 |
| NDUFB9 | B22 | DKFZp566O173 | FLJ22885 | LYRM3 | UQOR22 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa | Two-hybrid | BioGRID | 16169070 |
| PDCD6IP | AIP1 | Alix | DRIP4 | HP95 | MGC17003 | programmed cell death 6 interacting protein | - | HPRD | 12034747 |
| QTRT1 | FP3235 | TGT | queuine tRNA-ribosyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| RER1 | - | RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| RPL35A | - | ribosomal protein L35a | Two-hybrid | BioGRID | 16169070 |
| SH3D19 | EBP | EVE1 | Kryn | MGC105136 | MGC118910 | MGC118911 | MGC118912 | MGC118913 | SH3P19 | SH3 domain containing 19 | - | HPRD | 14551139 |
| SH3GL1 | CNSA1 | EEN | MGC111371 | SH3D2B | SH3P8 | SH3-domain GRB2-like 1 | Two-hybrid | BioGRID | 16169070 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | Endophilin A3 interacts with CIN85. | BIND | 11894096 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11894096 |
| SNRPN | DKFZp686C0927 | DKFZp686M12165 | DKFZp761I1912 | DKFZp762N022 | FLJ33569 | FLJ36996 | FLJ39265 | HCERN3 | MGC29886 | PWCR | RT-LI | SM-D | SMN | SNRNP-N | SNURF-SNRPN | small nuclear ribonucleoprotein polypeptide N | Two-hybrid | BioGRID | 16169070 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Two-hybrid | BioGRID | 16169070 |
| TOE1 | FLJ13949 | target of EGR1, member 1 (nuclear) | Two-hybrid | BioGRID | 16169070 |
| WDR33 | FLJ11294 | WDC146 | WD repeat domain 33 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 128 | 134 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-27 | 415 | 421 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-3p | 34 | 40 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-381 | 157 | 163 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.