Gene Page: SKI
Summary ?
| GeneID | 6497 |
| Symbol | SKI |
| Synonyms | SGS|SKV |
| Description | SKI proto-oncogene |
| Reference | MIM:164780|HGNC:HGNC:10896|Ensembl:ENSG00000157933|HPRD:01271|Vega:OTTHUMG00000001407 |
| Gene type | protein-coding |
| Map location | 1p36.33 |
| Pascal p-value | 0.025 |
| Fetal beta | 0.36 |
| DMG | 2 (# studies) |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10397932 | 1 | 2166155 | SKI | 2.33E-5 | -0.005 | 0.066 | DMG:Montano_2016 |
| cg21404476 | 1 | 2218549 | SKI | 2.133E-4 | 0.578 | 0.036 | DMG:Wockner_2014 |
| cg01938025 | 1 | 2228319 | SKI | 2.293E-4 | 0.456 | 0.036 | DMG:Wockner_2014 |
| cg19404444 | 1 | 2164602 | SKI | 3.227E-4 | 0.659 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
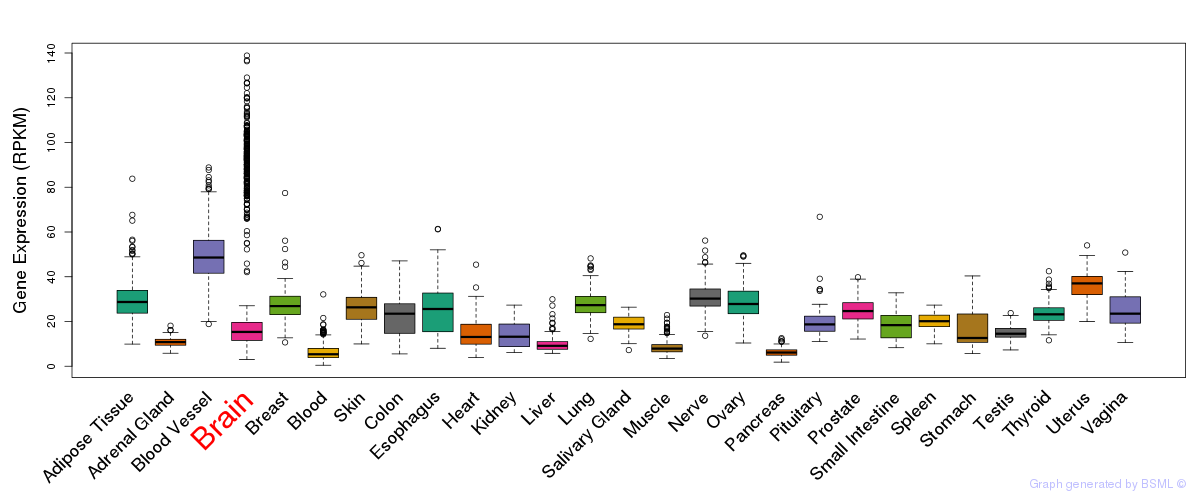
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTR | 0.85 | 0.85 |
| TSC1 | 0.82 | 0.83 |
| HERC2 | 0.81 | 0.82 |
| MACF1 | 0.81 | 0.82 |
| GOLGA3 | 0.81 | 0.81 |
| LRP1 | 0.81 | 0.80 |
| ZZEF1 | 0.80 | 0.81 |
| MLL2 | 0.80 | 0.79 |
| C14orf43 | 0.80 | 0.83 |
| MTF1 | 0.80 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.59 | -0.65 |
| UQCRB | -0.59 | -0.61 |
| RP11-884K10.1 | -0.58 | -0.63 |
| C1orf54 | -0.58 | -0.67 |
| GMFG | -0.57 | -0.65 |
| MTCP1NB | -0.57 | -0.60 |
| GNG11 | -0.56 | -0.64 |
| VAMP5 | -0.56 | -0.59 |
| AF347015.21 | -0.56 | -0.62 |
| RHOC | -0.55 | -0.58 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD,BioGRID | 12874272 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 12034730 |
| MECP2 | AUTSX3 | DKFZp686A24160 | MRX16 | MRX79 | MRXS13 | MRXSL | PPMX | RTS | RTT | methyl CpG binding protein 2 (Rett syndrome) | - | HPRD,BioGRID | 11441023 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Reconstituted Complex | BioGRID | 11430826 |
| NFIX | NF1A | nuclear factor I/X (CCAAT-binding transcription factor) | - | HPRD,BioGRID | 9380514 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 11430826 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Reconstituted Complex | BioGRID | 11430826 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 9927733 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | c-Ski homodimerizes with itself. | BIND | 10575014 |
| SKIL | SNO | SnoA | SnoN | SKI-like oncogene | - | HPRD,BioGRID | 9927733 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12874272 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | c-Ski interacts with Smad2. This interaction was modeled on a demonstrated interaction between Smad2 from an unspecified source and human c-Ski. | BIND | 10575014 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 10485843 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 12857746 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | c-Ski interacts with Smad3. This interaction was modeled on a demonstrated interaction between Smad3 from an unspecified source and human c-Ski. | BIND | 10575014 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD,BioGRID | 12419246 |12857746 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Smad4 interacts with Ski. This interaction was modeled on a demonstrated interaction between human Smad4 and Ski from an unspecified species. | BIND | 15761153 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | c-Ski interacts with Smad4. This interaction was modelled on a demonstrated interaction between Smad4 from an unspecified source and human c-Ski. | BIND | 10575014 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | Affinity Capture-Western Phenotypic Suppression Reconstituted Complex Two-hybrid | BioGRID | 9569025 |11278756 |11522815 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | - | HPRD | 9569025 |11522815 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | Ski interacts with alpha-tubulin. | BIND | 15806149 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY BMP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |