| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES
| 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT
| 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS
| 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS
| 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP
| 95 | 64 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP
| 157 | 104 | All SZGR 2.0 genes in this pathway |
| SILIGAN TARGETS OF EWS FLI1 FUSION DN
| 18 | 9 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP
| 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN
| 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP
| 197 | 135 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL
| 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC
| 108 | 74 | All SZGR 2.0 genes in this pathway |
| CEBALLOS TARGETS OF TP53 AND MYC DN
| 38 | 31 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4
| 261 | 153 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN 2FC DN
| 21 | 13 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN
| 584 | 395 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN
| 45 | 30 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN
| 200 | 112 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN
| 366 | 238 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN
| 68 | 44 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN
| 63 | 41 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR
| 544 | 307 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 UP
| 18 | 11 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN
| 1011 | 592 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN
| 191 | 123 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE
| 12 | 8 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION
| 30 | 21 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND RARA PLZF FUSION
| 22 | 14 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN
| 240 | 153 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12
| 317 | 177 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1
| 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN
| 253 | 192 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED
| 228 | 119 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP
| 857 | 456 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN
| 374 | 217 | All SZGR 2.0 genes in this pathway |
| ROLEF GLIS3 TARGETS
| 37 | 29 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
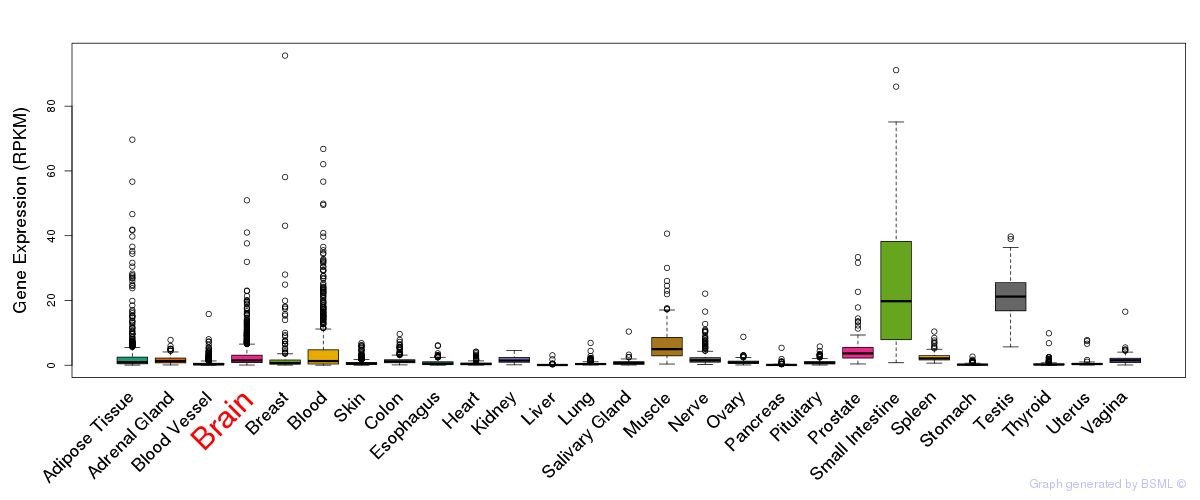
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation