Gene Page: SLC6A2
Summary ?
| GeneID | 6530 |
| Symbol | SLC6A2 |
| Synonyms | NAT1|NET|NET1|SLC6A5 |
| Description | solute carrier family 6 member 2 |
| Reference | MIM:163970|HGNC:HGNC:11048|Ensembl:ENSG00000103546|HPRD:01232|Vega:OTTHUMG00000133208 |
| Gene type | protein-coding |
| Map location | 16q12.2 |
| Pascal p-value | 0.439 |
| Fetal beta | 0.359 |
| eGene | Myers' cis & trans Meta |
| Support | NEUROTRANSMITTER METABOLISM |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16869851 | chr4 | 20690056 | SLC6A2 | 6530 | 0.12 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
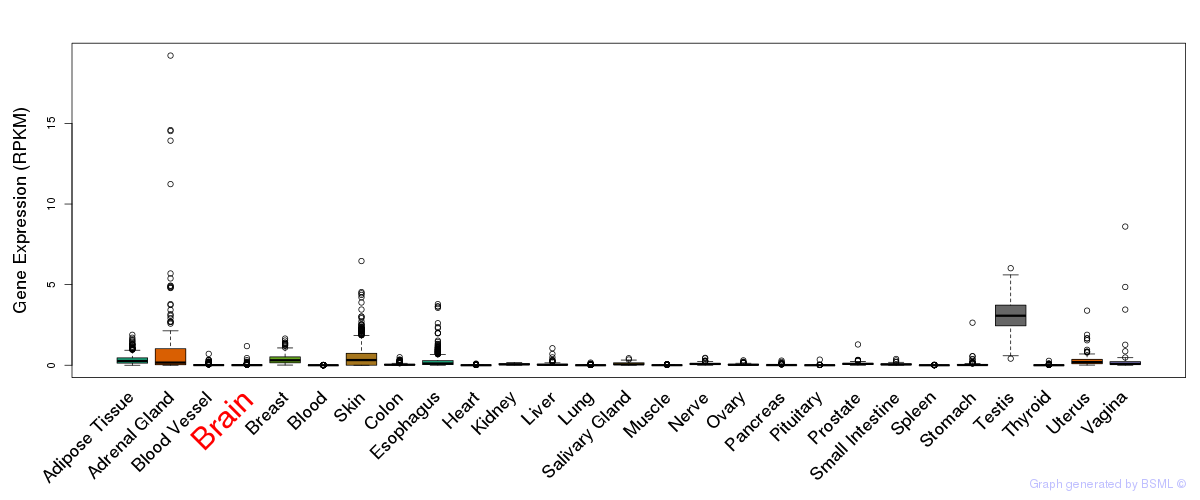
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CMTM6 | 0.74 | 0.70 |
| TMBIM6 | 0.73 | 0.58 |
| CYP2U1 | 0.71 | 0.65 |
| PTTG1IP | 0.70 | 0.53 |
| AP000872.1 | 0.70 | 0.62 |
| NXT2 | 0.70 | 0.46 |
| TRAM1 | 0.69 | 0.63 |
| ANO6 | 0.68 | 0.70 |
| SEPT2 | 0.68 | 0.55 |
| SLC35F5 | 0.67 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC098691.2 | -0.39 | -0.46 |
| AL022328.1 | -0.35 | -0.35 |
| AF347015.21 | -0.33 | -0.40 |
| FXYD1 | -0.32 | -0.37 |
| AC104794.1 | -0.32 | -0.30 |
| AC018755.7 | -0.32 | -0.34 |
| APOC1 | -0.30 | -0.29 |
| S100A13 | -0.29 | -0.34 |
| MYL3 | -0.29 | -0.34 |
| AF347015.18 | -0.29 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005334 | norephinephrine:sodium symporter activity | TAS | Neurotransmitter (GO term level: 10) | 2008212 |
| GO:0005328 | neurotransmitter:sodium symporter activity | IEA | Neurotransmitter (GO term level: 9) | - |
| GO:0005515 | protein binding | IPI | 11343649 | |
| GO:0008504 | monoamine transmembrane transporter activity | IDA | 16024787 | |
| GO:0015293 | symporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 2008212 |
| GO:0006836 | neurotransmitter transport | IEA | neuron, Neurotransmitter (GO term level: 5) | - |
| GO:0015844 | monoamine transport | IDA | 16024787 | |
| GO:0048265 | response to pain | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 2008212 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005739 | mitochondrion | IDA | 18029348 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 2008212 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE COMPOUND SLC TRANSPORTERS | 27 | 16 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| YAMASHITA METHYLATED IN PROSTATE CANCER | 57 | 29 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 UP | 65 | 43 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |