Gene Page: SLC6A3
Summary ?
| GeneID | 6531 |
| Symbol | SLC6A3 |
| Synonyms | DAT|DAT1|PKDYS |
| Description | solute carrier family 6 member 3 |
| Reference | MIM:126455|HGNC:HGNC:11049|Ensembl:ENSG00000142319|HPRD:00543|Vega:OTTHUMG00000131016 |
| Gene type | protein-coding |
| Map location | 5p15.3 |
| Pascal p-value | 0.876 |
| Sherlock p-value | 0.925 |
| DMG | 1 (# studies) |
| Support | DOPAMINE NEUROTRANSMITTER METABOLISM |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3854 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26205131 | 5 | 1444878 | SLC6A3 | -0.021 | 0.29 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
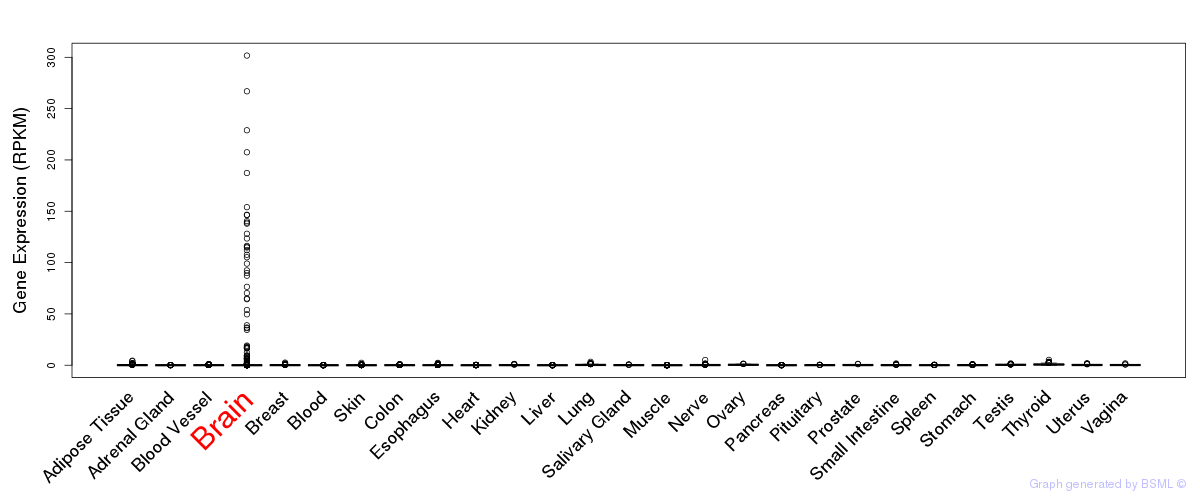
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARSB | 0.92 | 0.93 |
| PTPN9 | 0.91 | 0.94 |
| TSPAN14 | 0.90 | 0.91 |
| DTX4 | 0.90 | 0.93 |
| CDK5R1 | 0.90 | 0.92 |
| PACS1 | 0.90 | 0.92 |
| SARM1 | 0.90 | 0.93 |
| SAP130 | 0.89 | 0.92 |
| DYRK2 | 0.89 | 0.94 |
| GPRIN1 | 0.89 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| S100B | -0.73 | -0.84 |
| MT-CO2 | -0.73 | -0.86 |
| C5orf53 | -0.73 | -0.78 |
| AF347015.33 | -0.72 | -0.84 |
| AF347015.31 | -0.72 | -0.84 |
| FXYD1 | -0.72 | -0.83 |
| AF347015.27 | -0.71 | -0.83 |
| MT-CYB | -0.69 | -0.81 |
| AF347015.8 | -0.69 | -0.84 |
| AC021016.1 | -0.69 | -0.81 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005330 | dopamine:sodium symporter activity | IEA | Neurotransmitter, dopamine (GO term level: 10) | - |
| GO:0005515 | protein binding | IPI | 11343649 | |
| GO:0015293 | symporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0015872 | dopamine transport | IEA | Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0042053 | regulation of dopamine metabolic process | IEA | dopamine (GO term level: 9) | - |
| GO:0042420 | dopamine catabolic process | IEA | Neurotransmitter, dopamine (GO term level: 9) | - |
| GO:0042416 | dopamine biosynthetic process | IEA | Neurotransmitter, dopamine (GO term level: 9) | - |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0007608 | sensory perception of smell | IEA | - | |
| GO:0007595 | lactation | IEA | - | |
| GO:0015844 | monoamine transport | IDA | 16024787 | |
| GO:0021984 | adenohypophysis development | IEA | - | |
| GO:0040018 | positive regulation of multicellular organism growth | IEA | - | |
| GO:0060134 | prepulse inhibition | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 10823899 | |
| GO:0005886 | plasma membrane | IDA | 11343649 | |
| GO:0005887 | integral to plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE COMPOUND SLC TRANSPORTERS | 27 | 16 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 5P15 AMPLICON | 26 | 15 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |