Gene Page: BMP6
Summary ?
| GeneID | 654 |
| Symbol | BMP6 |
| Synonyms | VGR|VGR1 |
| Description | bone morphogenetic protein 6 |
| Reference | MIM:112266|HGNC:HGNC:1073|Ensembl:ENSG00000153162|HPRD:00211|Vega:OTTHUMG00000014217 |
| Gene type | protein-coding |
| Map location | 6p24-p23 |
| Pascal p-value | 0.11 |
| Sherlock p-value | 0.66 |
| Fetal beta | -0.136 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs546975 | chr1 | 91963738 | BMP6 | 654 | 0.03 | trans | ||
| rs4236095 | chr6 | 47368686 | BMP6 | 654 | 0.02 | trans | ||
| snp_a-2266657 | 0 | BMP6 | 654 | 0.15 | trans | |||
| rs2430483 | chr7 | 104548631 | BMP6 | 654 | 0.07 | trans | ||
| rs2061799 | chr9 | 97795003 | BMP6 | 654 | 0.01 | trans | ||
| rs10149864 | chr14 | 86719683 | BMP6 | 654 | 0 | trans | ||
| rs5029382 | chr18 | 45981525 | BMP6 | 654 | 0.11 | trans | ||
| rs1877412 | chr18 | 45987674 | BMP6 | 654 | 0.09 | trans | ||
| rs11873703 | chr18 | 61448702 | BMP6 | 654 | 0.19 | trans | ||
| rs16999569 | chr19 | 22902709 | BMP6 | 654 | 0.09 | trans | ||
| rs12973782 | chr19 | 22905126 | BMP6 | 654 | 0.09 | trans | ||
| snp_a-2090370 | 0 | BMP6 | 654 | 0.09 | trans | |||
| rs12980129 | chr19 | 22908910 | BMP6 | 654 | 0.02 | trans | ||
| rs2194111 | chr19 | 22922548 | BMP6 | 654 | 0.09 | trans | ||
| rs6095741 | chr20 | 48666589 | BMP6 | 654 | 0.02 | trans |
Section II. Transcriptome annotation
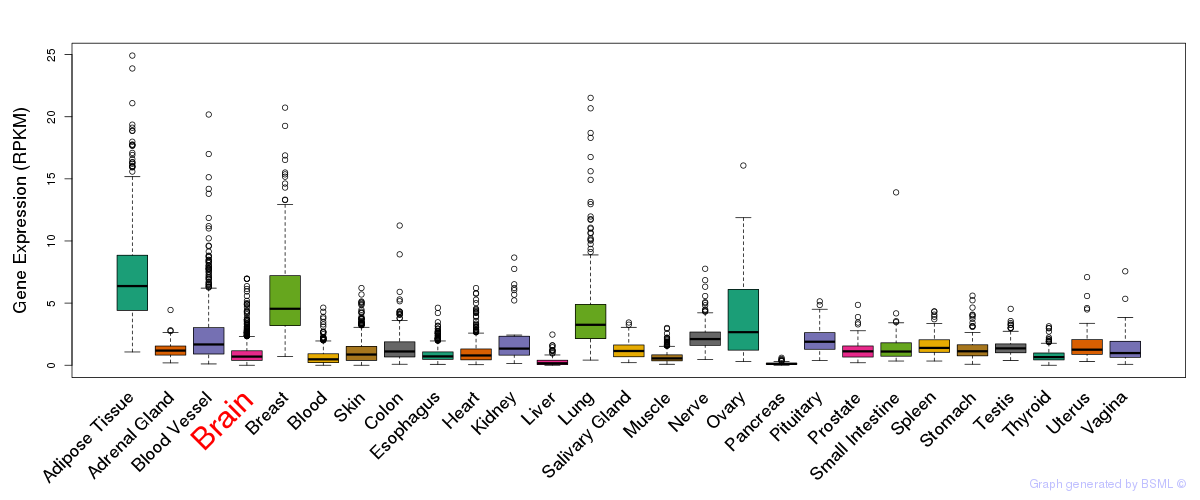
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC4A4 | 0.86 | 0.79 |
| PPAP2B | 0.86 | 0.85 |
| ATP13A4 | 0.85 | 0.77 |
| ACSS3 | 0.84 | 0.81 |
| AQP4 | 0.83 | 0.77 |
| SSPN | 0.83 | 0.75 |
| SLC1A3 | 0.83 | 0.84 |
| PDLIM5 | 0.82 | 0.80 |
| ITPR2 | 0.82 | 0.73 |
| ADD3 | 0.82 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.36 | -0.39 |
| AC011491.1 | -0.35 | -0.37 |
| RAB33A | -0.34 | -0.37 |
| DCTPP1 | -0.33 | -0.36 |
| DPF1 | -0.33 | -0.30 |
| BZW2 | -0.32 | -0.33 |
| GPR21 | -0.32 | -0.29 |
| POLB | -0.32 | -0.34 |
| ACTL6B | -0.31 | -0.31 |
| ARL4D | -0.31 | -0.29 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006955 | immune response | IMP | 16886151 | |
| GO:0006954 | inflammatory response | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030501 | positive regulation of bone mineralization | IDA | 18436533 | |
| GO:0030509 | BMP signaling pathway | IEA | - | |
| GO:0051216 | cartilage development | IEA | - | |
| GO:0032349 | positive regulation of aldosterone biosynthetic process | IDA | 16527843 | |
| GO:0040007 | growth | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 16527843 | |
| GO:0045669 | positive regulation of osteoblast differentiation | IDA | 18436533 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE | 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 55 | 62 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 55 | 61 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-219 | 1015 | 1021 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.