Gene Page: BMP7
Summary ?
| GeneID | 655 |
| Symbol | BMP7 |
| Synonyms | OP-1 |
| Description | bone morphogenetic protein 7 |
| Reference | MIM:112267|HGNC:HGNC:1074|Ensembl:ENSG00000101144|HPRD:00212|Vega:OTTHUMG00000032812 |
| Gene type | protein-coding |
| Map location | 20q13 |
| Pascal p-value | 0.596 |
| Fetal beta | 0.281 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| Expression | Meta-analysis of gene expression | P value: 1.368 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23420286 | 20 | 55835350 | BMP7 | 1.85E-4 | 0.65 | 0.034 | DMG:Wockner_2014 |
| cg15069906 | 20 | 55841888 | BMP7 | 3.458E-4 | -0.285 | 0.041 | DMG:Wockner_2014 |
| cg20292547 | 20 | 55841342 | BMP7 | 4.364E-4 | -0.2 | 0.045 | DMG:Wockner_2014 |
| cg00340850 | 20 | 55500523 | BMP7 | 1.29E-10 | -0.027 | 4.83E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6849842 | chr4 | 185436254 | BMP7 | 655 | 0.03 | trans |
Section II. Transcriptome annotation
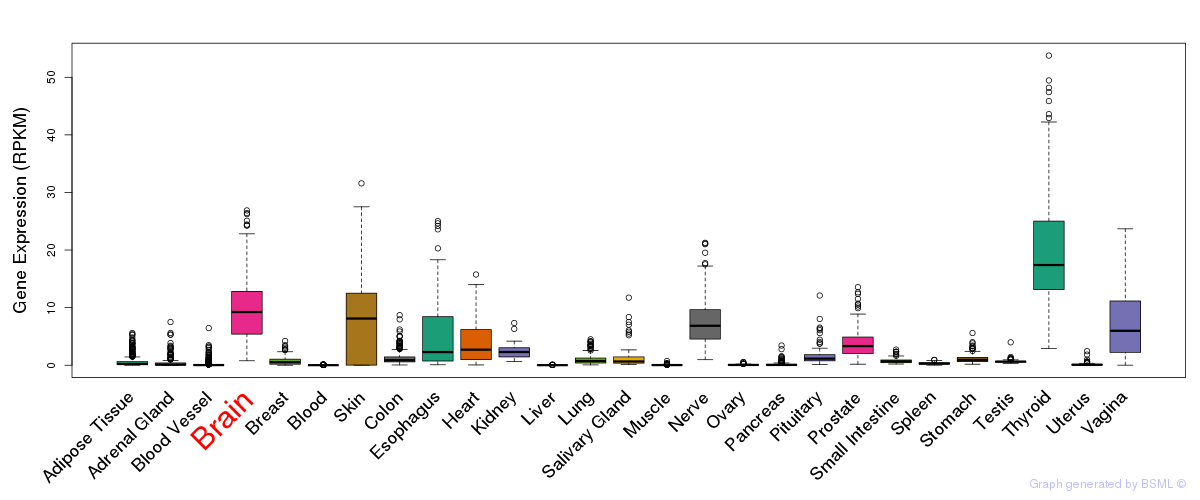
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1219 | 0.95 | 0.97 |
| TAOK1 | 0.94 | 0.96 |
| BRAF | 0.94 | 0.96 |
| VCPIP1 | 0.94 | 0.96 |
| ZYG11B | 0.94 | 0.96 |
| RLIM | 0.93 | 0.96 |
| DENND5B | 0.93 | 0.96 |
| NUFIP2 | 0.93 | 0.96 |
| SEL1L | 0.92 | 0.94 |
| ZBTB44 | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.66 | -0.75 |
| HIGD1B | -0.62 | -0.74 |
| TLCD1 | -0.62 | -0.70 |
| C19orf36 | -0.62 | -0.67 |
| S100A16 | -0.61 | -0.70 |
| CST3 | -0.60 | -0.70 |
| AC021016.1 | -0.60 | -0.68 |
| MT-CO2 | -0.60 | -0.72 |
| SERPINB6 | -0.60 | -0.66 |
| ENHO | -0.60 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 12478285 |12667445 | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0001503 | ossification | IEA | - | |
| GO:0001707 | mesoderm formation | IEA | - | |
| GO:0001837 | epithelial to mesenchymal transition | TAS | 14679171 | |
| GO:0001822 | kidney development | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0048468 | cell development | IEA | - | |
| GO:0007435 | salivary gland morphogenesis | IEA | - | |
| GO:0007389 | pattern specification process | IEA | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0030501 | positive regulation of bone mineralization | IDA | 18436533 | |
| GO:0030509 | BMP signaling pathway | IEA | - | |
| GO:0051216 | cartilage development | IEA | - | |
| GO:0048754 | branching morphogenesis of a tube | IEA | - | |
| GO:0040007 | growth | IEA | - | |
| GO:0045941 | positive regulation of transcription | IDA | 14517293 | |
| GO:0045786 | negative regulation of cell cycle | IDA | 11502704 | |
| GO:0045669 | positive regulation of osteoblast differentiation | IDA | 18436533 | |
| GO:0048593 | camera-type eye morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1 | ACTRI | ACVR1A | ACVRLK2 | ALK2 | FOP | SKR1 | TSRI | activin A receptor, type I | - | HPRD,BioGRID | 8006002 |
| ACVR2A | ACTRII | ACVR2 | activin A receptor, type IIA | - | HPRD,BioGRID | 9748228 |
| ACVR2B | ACTRIIB | ActR-IIB | MGC116908 | activin A receptor, type IIB | - | HPRD,BioGRID | 9748228 |
| BMP7 | OP-1 | bone morphogenetic protein 7 | Co-purification | BioGRID | 12478285 |
| BMPR1A | 10q23del | ACVRLK3 | ALK3 | CD292 | bone morphogenetic protein receptor, type IA | Reconstituted Complex | BioGRID | 8006002 |
| BMPR1A | 10q23del | ACVRLK3 | ALK3 | CD292 | bone morphogenetic protein receptor, type IA | - | HPRD | 7791754 |8605097 |
| BMPR1B | ALK-6 | ALK6 | CDw293 | bone morphogenetic protein receptor, type IB | - | HPRD | 8605097 |
| BMPR1B | ALK-6 | ALK6 | CDw293 | bone morphogenetic protein receptor, type IB | Reconstituted Complex | BioGRID | 8006002 |
| BMPR2 | BMPR-II | BMPR3 | BMR2 | BRK-3 | FLJ41585 | FLJ76945 | PPH1 | T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) | - | HPRD,BioGRID | 7791754 |
| ENG | CD105 | END | FLJ41744 | HHT1 | ORW | ORW1 | endoglin | - | HPRD,BioGRID | 9872992 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD | 7791754 |
| NOG | SYM1 | SYNS1 | noggin | - | HPRD,BioGRID | 12478285 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Phenotypic Suppression | BioGRID | 11438941 |
| SOSTDC1 | CDA019 | DKFZp564D206 | ECTODIN | USAG1 | sclerostin domain containing 1 | - | HPRD,BioGRID | 14623234 |15020244 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID ALK2 PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM3 | 70 | 37 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST VS SYNOVIAL SARCOMA UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS UP | 10 | 8 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA CNN1 DN | 20 | 18 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| MALIK REPRESSED BY ESTROGEN | 12 | 10 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-542-3p | 438 | 444 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.