Gene Page: SNAPC3
Summary ?
| GeneID | 6619 |
| Symbol | SNAPC3 |
| Synonyms | PTFbeta|SNAP50 |
| Description | small nuclear RNA activating complex polypeptide 3 |
| Reference | MIM:602348|HGNC:HGNC:11136|HPRD:03826| |
| Gene type | protein-coding |
| Map location | 9p22.3 |
| Pascal p-value | 1.049E-4 |
| Sherlock p-value | 0.247 |
| Fetal beta | 1.213 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
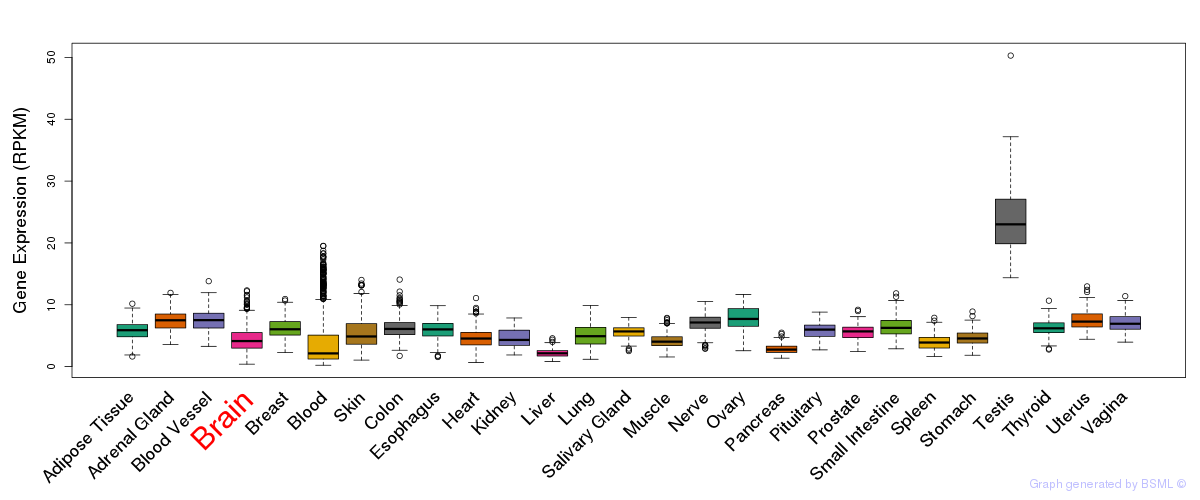
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NFYA | 0.94 | 0.80 |
| IGSF3 | 0.94 | 0.90 |
| BCL11B | 0.90 | 0.83 |
| USP6NL | 0.90 | 0.88 |
| EPHB1 | 0.90 | 0.85 |
| TBR1 | 0.90 | 0.72 |
| VANGL2 | 0.90 | 0.87 |
| AC087698.1 | 0.89 | 0.75 |
| IGFBPL1 | 0.89 | 0.89 |
| SOX11 | 0.89 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNFSF12 | -0.59 | -0.65 |
| C5orf53 | -0.57 | -0.85 |
| FBXO2 | -0.57 | -0.68 |
| PTH1R | -0.57 | -0.69 |
| HLA-F | -0.57 | -0.82 |
| TMEM54 | -0.56 | -0.61 |
| C19orf66 | -0.56 | -0.48 |
| LHPP | -0.56 | -0.64 |
| SERPINB6 | -0.56 | -0.71 |
| PPAP2A | -0.56 | -0.62 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 24HR UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR UP | 9 | 5 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |