Gene Page: SNRPB
Summary ?
| GeneID | 6628 |
| Symbol | SNRPB |
| Synonyms | CCMS|COD|SNRPB1|Sm-B/B'|SmB/B'|SmB/SmB'|snRNP-B |
| Description | small nuclear ribonucleoprotein polypeptides B and B1 |
| Reference | MIM:182282|HGNC:HGNC:11153|Ensembl:ENSG00000125835|HPRD:01655|Vega:OTTHUMG00000031694 |
| Gene type | protein-coding |
| Map location | 20p13 |
| Pascal p-value | 0.061 |
| Sherlock p-value | 0.053 |
| Fetal beta | 0.7 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04435241 | 20 | 2442475 | SNRPB | 1.779E-4 | 0.268 | 0.034 | DMG:Wockner_2014 |
| cg11521799 | 20 | 2451663 | SNRPB | 4.71E-8 | -0.015 | 1.27E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
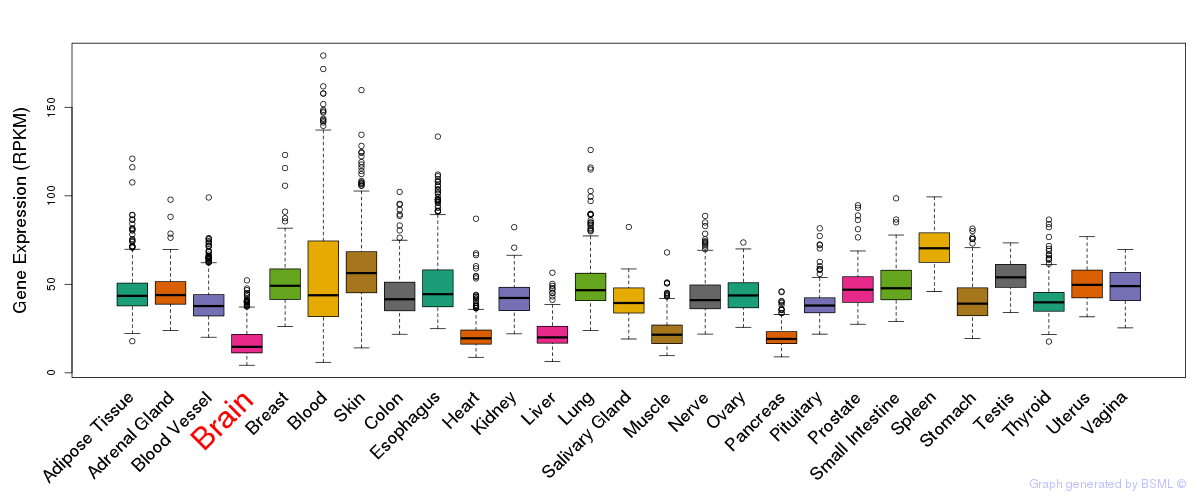
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DPP9 | 0.86 | 0.88 |
| ZNF687 | 0.86 | 0.86 |
| ATG2A | 0.84 | 0.88 |
| NUP210 | 0.84 | 0.87 |
| MED15 | 0.84 | 0.86 |
| POLG | 0.84 | 0.85 |
| FOXRED2 | 0.83 | 0.86 |
| RPAP1 | 0.83 | 0.87 |
| CORO2B | 0.83 | 0.84 |
| UPF1 | 0.83 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.63 | -0.66 |
| SYCP3 | -0.61 | -0.70 |
| C1orf54 | -0.60 | -0.65 |
| GNG11 | -0.58 | -0.62 |
| AF347015.31 | -0.57 | -0.55 |
| MT-CO2 | -0.56 | -0.55 |
| AL050337.1 | -0.56 | -0.63 |
| CLEC2B | -0.55 | -0.63 |
| HIGD1B | -0.54 | -0.53 |
| NOSTRIN | -0.53 | -0.51 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| CLNS1A | CLCI | CLNS1B | ICln | chloride channel, nucleotide-sensitive, 1A | Affinity Capture-Western | BioGRID | 11713266 |
| COIL | CLN80 | p80-coilin | coilin | - | HPRD,BioGRID | 11641277 |
| CTDP1 | CCFDN | FCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 | - | HPRD | 12560496 |
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | - | HPRD,BioGRID | 10601333 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD | 11149922 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EFTUD2 | DKFZp686E24196 | FLJ44695 | KIAA0031 | Snrp116 | Snu114 | U5-116KD | elongation factor Tu GTP binding domain containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| FNBP4 | DKFZp686M14102 | DKFZp779I1064 | FBP30 | FLJ41904 | KIAA1014 | formin binding protein 4 | Protein-peptide | BioGRID | 11604498 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | - | HPRD | 10725331 |
| GEMIN5 | DKFZp586M1824 | MGC142174 | gem (nuclear organelle) associated protein 5 | - | HPRD | 11714716 |
| GEMIN6 | FLJ23459 | gem (nuclear organelle) associated protein 6 | Gemin6 interacts with SmB. | BIND | 11748230 |
| GEMIN6 | FLJ23459 | gem (nuclear organelle) associated protein 6 | Gemin6 interacts with SNRPB (SmB) | BIND | 15939020 |
| GEMIN7 | SIP3 | gem (nuclear organelle) associated protein 7 | GEMIN7 interacts with SNRPB (SmB) | BIND | 12065586 |
| HTATSF1 | TAT-SF1 | dJ196E23.2 | HIV-1 Tat specific factor 1 | Affinity Capture-MS | BioGRID | 17353931 |
| KHK | - | ketohexokinase (fructokinase) | Affinity Capture-MS | BioGRID | 17353931 |
| LGALS1 | DKFZp686E23103 | GAL1 | GBP | lectin, galactoside-binding, soluble, 1 | Affinity Capture-Western | BioGRID | 11522829 |
| LSM11 | FLJ38273 | LSM11, U7 small nuclear RNA associated | - | HPRD | 12975319 |
| PRPF8 | HPRP8 | PRP8 | PRPC8 | RP13 | PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| PSMA3 | HC8 | MGC12306 | MGC32631 | PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 | Two-hybrid | BioGRID | 16169070 |
| RBBP6 | DKFZp686P0638 | DKFZp761B2423 | MY038 | P2P-R | PACT | RBQ-1 | SNAMA | retinoblastoma binding protein 6 | Sm interacts with PACT. | BIND | 9010216 |
| SART3 | DSAP1 | KIAA0156 | MGC138188 | P100 | RP11-13G14 | TIP110 | p110 | p110(nrb) | squamous cell carcinoma antigen recognized by T cells 3 | Affinity Capture-MS | BioGRID | 17353931 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | SMN interacts with SmB'. | BIND | 15494309 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD | 11720283 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD,BioGRID | 9323129|11720283 |
| SMN2 | BCD541 | C-BCD541 | FLJ76644 | MGC20996 | MGC5208 | SMNC | survival of motor neuron 2, centromeric | - | HPRD | 9323129 |
| SNRNP200 | ASCC3L1 | BRR2 | HELIC2 | U5-200KD | small nuclear ribonucleoprotein 200kDa (U5) | Affinity Capture-MS | BioGRID | 17353931 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | - | HPRD,BioGRID | 10025403 |
| SNUPN | KPNBL | RNUT1 | Snurportin1 | snurportin 1 | Reconstituted Complex | BioGRID | 12095920 |
| STXBP2 | Hunc18b | MUNC18-2 | UNC18-2 | UNC18B | pp10122 | syntaxin binding protein 2 | - | HPRD | 11748230 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | - | HPRD | 12065586 |
| TOP3B | FLJ39376 | topoisomerase (DNA) III beta | - | HPRD,BioGRID | 11549288 |
| WBP4 | FBP21 | MGC117310 | WW domain binding protein 4 (formin binding protein 21) | - | HPRD | 9724750 |
| WBP4 | FBP21 | MGC117310 | WW domain binding protein 4 (formin binding protein 21) | Protein-peptide | BioGRID | 11604498 |
| WDR77 | HKMT1069 | MEP50 | MGC2722 | Nbla10071 | RP11-552M11.3 | p44 | WD repeat domain 77 | - | HPRD,BioGRID | 11756452 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | 23 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE UP | 47 | 32 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L1 G1 DN | 12 | 6 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA UP | 45 | 30 | All SZGR 2.0 genes in this pathway |
| NADLER HYPERGLYCEMIA AT OBESITY | 58 | 35 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP | 79 | 40 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 UP | 45 | 21 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |