Gene Page: SOD2
Summary ?
| GeneID | 6648 |
| Symbol | SOD2 |
| Synonyms | IPOB|MNSOD|MVCD6 |
| Description | superoxide dismutase 2, mitochondrial |
| Reference | MIM:147460|HGNC:HGNC:11180|Ensembl:ENSG00000112096|HPRD:00938|Vega:OTTHUMG00000015940 |
| Gene type | protein-coding |
| Map location | 6q25.3 |
| Pascal p-value | 0.091 |
| Sherlock p-value | 0.86 |
| Fetal beta | -1.79 |
| eGene | Myers' cis & trans Meta |
| Support | G2Cdb.human_clathrin G2Cdb.human_mitochondria G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2820477 | chr1 | 68708633 | SOD2 | 6648 | 0.09 | trans | ||
| rs473019 | chr1 | 72296394 | SOD2 | 6648 | 0.12 | trans | ||
| rs1342707 | chr1 | 147264296 | SOD2 | 6648 | 0.17 | trans | ||
| rs12076447 | chr1 | 159552549 | SOD2 | 6648 | 0.05 | trans | ||
| rs10494571 | chr1 | 183930700 | SOD2 | 6648 | 0.15 | trans | ||
| rs6660906 | chr1 | 227031843 | SOD2 | 6648 | 0.17 | trans | ||
| rs17541094 | chr2 | 66266798 | SOD2 | 6648 | 0.07 | trans | ||
| rs1196156 | chr2 | 182924287 | SOD2 | 6648 | 0.04 | trans | ||
| rs16847252 | chr3 | 163481290 | SOD2 | 6648 | 1.165E-4 | trans | ||
| rs16846456 | chr3 | 172757337 | SOD2 | 6648 | 5.684E-4 | trans | ||
| rs17237063 | chr3 | 182134513 | SOD2 | 6648 | 0.01 | trans | ||
| rs591585 | chr4 | 43424979 | SOD2 | 6648 | 0.04 | trans | ||
| rs16882559 | chr5 | 9362994 | SOD2 | 6648 | 0.19 | trans | ||
| rs9293020 | chr5 | 22520547 | SOD2 | 6648 | 0.18 | trans | ||
| rs888819 | chr5 | 80257108 | SOD2 | 6648 | 0.01 | trans | ||
| rs10515260 | chr5 | 97076547 | SOD2 | 6648 | 3.191E-6 | trans | ||
| rs10515267 | chr5 | 100128201 | SOD2 | 6648 | 0.08 | trans | ||
| rs16884319 | chr6 | 20960006 | SOD2 | 6648 | 0.18 | trans | ||
| rs16873760 | chr6 | 45729388 | SOD2 | 6648 | 0.02 | trans | ||
| rs16873861 | chr6 | 45752854 | SOD2 | 6648 | 0.02 | trans | ||
| rs17156205 | chr7 | 28060204 | SOD2 | 6648 | 0.17 | trans | ||
| rs6946007 | chr7 | 28061329 | SOD2 | 6648 | 2.048E-4 | trans | ||
| rs6956395 | chr7 | 28072244 | SOD2 | 6648 | 0 | trans | ||
| snp_a-2079112 | 0 | SOD2 | 6648 | 0.17 | trans | |||
| rs7790094 | chr7 | 152786528 | SOD2 | 6648 | 0.18 | trans | ||
| rs735485 | chr8 | 3706731 | SOD2 | 6648 | 0.05 | trans | ||
| rs13282254 | chr8 | 126625316 | SOD2 | 6648 | 0.14 | trans | ||
| rs6476030 | chr9 | 289060 | SOD2 | 6648 | 0.03 | trans | ||
| rs7848093 | chr9 | 5875200 | SOD2 | 6648 | 0.01 | trans | ||
| rs10116858 | chr9 | 20300988 | SOD2 | 6648 | 1.175E-4 | trans | ||
| rs17742994 | chr9 | 115555409 | SOD2 | 6648 | 0.12 | trans | ||
| rs17376956 | chr10 | 11179948 | SOD2 | 6648 | 0.15 | trans | ||
| rs11254107 | chr10 | 16650659 | SOD2 | 6648 | 0.18 | trans | ||
| rs1061577 | chr10 | 28101310 | SOD2 | 6648 | 0.15 | trans | ||
| rs3802522 | chr10 | 28341865 | SOD2 | 6648 | 2.705E-7 | trans | ||
| rs10508730 | chr10 | 28403391 | SOD2 | 6648 | 2.705E-7 | trans | ||
| rs16936983 | chr10 | 37871050 | SOD2 | 6648 | 0.03 | trans | ||
| rs1344342 | chr10 | 37958911 | SOD2 | 6648 | 0.03 | trans | ||
| rs1108733 | chr10 | 37993286 | SOD2 | 6648 | 0.03 | trans | ||
| rs7080407 | chr10 | 54168712 | SOD2 | 6648 | 0.01 | trans | ||
| rs7089703 | chr10 | 119182182 | SOD2 | 6648 | 0.07 | trans | ||
| rs10743139 | chr11 | 10425326 | SOD2 | 6648 | 4.312E-4 | trans | ||
| snp_a-2125340 | 0 | SOD2 | 6648 | 0.15 | trans | |||
| rs2512874 | chr11 | 131461291 | SOD2 | 6648 | 0.15 | trans | ||
| rs11172848 | chr12 | 40007955 | SOD2 | 6648 | 0.09 | trans | ||
| rs1921012 | chr12 | 67176791 | SOD2 | 6648 | 0.05 | trans | ||
| rs4762419 | chr12 | 95771835 | SOD2 | 6648 | 0.12 | trans | ||
| rs9645995 | chr13 | 47246048 | SOD2 | 6648 | 0 | trans | ||
| rs9598109 | chr13 | 60878618 | SOD2 | 6648 | 0.03 | trans | ||
| rs8012787 | chr14 | 36085745 | SOD2 | 6648 | 0.14 | trans | ||
| snp_a-2138420 | 0 | SOD2 | 6648 | 0.14 | trans | |||
| rs11623645 | chr14 | 36195311 | SOD2 | 6648 | 0.07 | trans | ||
| rs9989174 | chr14 | 82663493 | SOD2 | 6648 | 0.15 | trans | ||
| rs1958669 | chr14 | 82675761 | SOD2 | 6648 | 0.01 | trans | ||
| rs962253 | chr15 | 99339832 | SOD2 | 6648 | 0.2 | trans | ||
| rs2684763 | chr15 | 99365485 | SOD2 | 6648 | 0.2 | trans | ||
| rs16971043 | chr16 | 72604104 | SOD2 | 6648 | 0 | trans | ||
| rs4303487 | chr16 | 90033254 | SOD2 | 6648 | 0.02 | trans | ||
| rs16968345 | chr17 | 73948230 | SOD2 | 6648 | 0.02 | trans | ||
| rs2448755 | chr18 | 65344801 | SOD2 | 6648 | 0.01 | trans | ||
| rs965018 | chr20 | 41531585 | SOD2 | 6648 | 0.1 | trans | ||
| rs1206765 | chr20 | 45586718 | SOD2 | 6648 | 0.18 | trans | ||
| rs734600 | chr20 | 45591681 | SOD2 | 6648 | 0.18 | trans | ||
| rs17342476 | chrX | 102243644 | SOD2 | 6648 | 0 | trans | ||
| rs17342483 | chrX | 102250155 | SOD2 | 6648 | 0.01 | trans | ||
| rs17342504 | chrX | 102272107 | SOD2 | 6648 | 0 | trans | ||
| snp_a-2138851 | 0 | SOD2 | 6648 | 0 | trans | |||
| snp_a-2059928 | 0 | SOD2 | 6648 | 0 | trans |
Section II. Transcriptome annotation
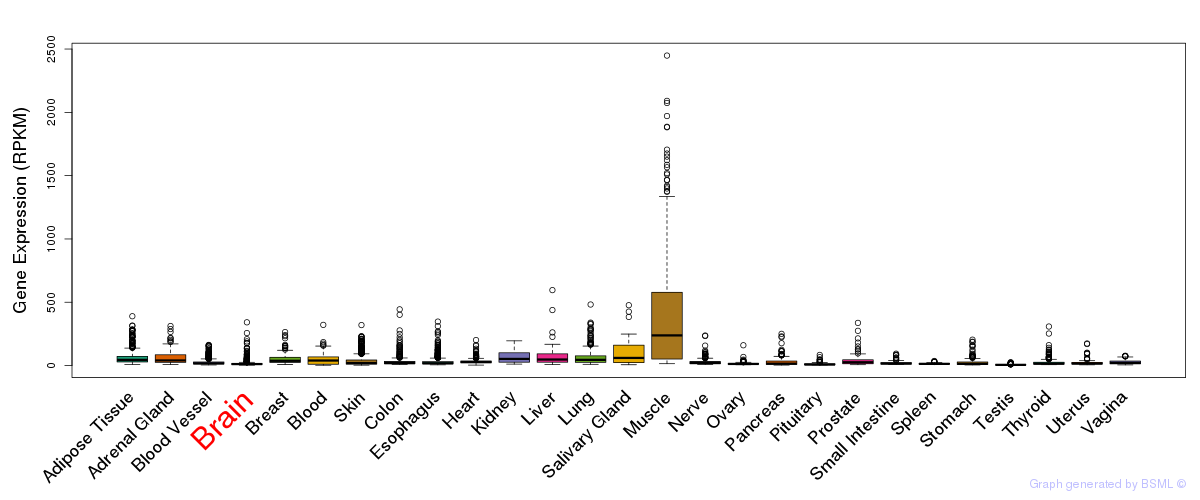
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C3 | 0.81 | 0.77 |
| CD37 | 0.81 | 0.77 |
| C1QC | 0.80 | 0.81 |
| APBB1IP | 0.80 | 0.77 |
| LILRB4 | 0.78 | 0.78 |
| PTPN6 | 0.77 | 0.76 |
| TBXAS1 | 0.77 | 0.75 |
| ITGB2 | 0.77 | 0.69 |
| HCLS1 | 0.76 | 0.76 |
| ADORA3 | 0.75 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC004017.1 | -0.39 | -0.41 |
| TTLL2 | -0.39 | -0.37 |
| ZNF124 | -0.39 | -0.37 |
| ZNF551 | -0.39 | -0.38 |
| GJC1 | -0.39 | -0.38 |
| SATB2 | -0.39 | -0.33 |
| TMEM57 | -0.38 | -0.36 |
| TBC1D30 | -0.38 | -0.35 |
| ZNF286 | -0.38 | -0.38 |
| MLLT3 | -0.38 | -0.34 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004784 | superoxide dismutase activity | IDA | 14980699 | |
| GO:0004784 | superoxide dismutase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000303 | response to superoxide | IEA | - | |
| GO:0000303 | response to superoxide | TAS | 9462746 | |
| GO:0001315 | age-dependent response to reactive oxygen species | IMP | 14980699 | |
| GO:0001315 | age-dependent response to reactive oxygen species | ISS | - | |
| GO:0001836 | release of cytochrome c from mitochondria | IEA | - | |
| GO:0006302 | double-strand break repair | IEA | - | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | IMP | 9393747 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | ISS | - | |
| GO:0006801 | superoxide metabolic process | IDA | 14980699 | |
| GO:0006801 | superoxide metabolic process | IEA | - | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0006979 | response to oxidative stress | TAS | 9462746 | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0042542 | response to hydrogen peroxide | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 15087454 | |
| GO:0005743 | mitochondrial inner membrane | IEA | - | |
| GO:0005759 | mitochondrial matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LONGEVITY PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP | 52 | 38 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE UP | 52 | 36 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS CML DIVIDING UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN RESPONSE TO MP470 DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN RESPONSE TO MP470 UP | 20 | 11 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 UP | 64 | 45 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA DN | 41 | 33 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| MURATA VIRULENCE OF H PILORI | 24 | 16 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA | 46 | 32 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| FERRANDO LYL1 NEIGHBORS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS UP | 48 | 36 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 4HR | 54 | 36 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| MOOTHA ROS | 7 | 7 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| MARTINELLI IMMATURE NEUTROPHIL DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING UP | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP | 55 | 41 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |