Gene Page: SPOCK1
Summary ?
| GeneID | 6695 |
| Symbol | SPOCK1 |
| Synonyms | SPOCK|TESTICAN|TIC1 |
| Description | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| Reference | MIM:602264|HGNC:HGNC:11251|Ensembl:ENSG00000152377|HPRD:11887|Vega:OTTHUMG00000129157 |
| Gene type | protein-coding |
| Map location | 5q31.2 |
| Pascal p-value | 0.007 |
| Sherlock p-value | 0.006 |
| Fetal beta | -0.707 |
| eGene | Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17600276 | 5 | 136433386 | SPOCK1 | ENSG00000152377.8 | 2.66E-6 | 0.01 | 500682 | gtex_brain_putamen_basal |
| rs201543316 | 5 | 136436560 | SPOCK1 | ENSG00000152377.8 | 3.473E-6 | 0.01 | 497508 | gtex_brain_putamen_basal |
| rs965166 | 5 | 136439130 | SPOCK1 | ENSG00000152377.8 | 1.641E-6 | 0.01 | 494938 | gtex_brain_putamen_basal |
| rs965167 | 5 | 136439139 | SPOCK1 | ENSG00000152377.8 | 1.664E-6 | 0.01 | 494929 | gtex_brain_putamen_basal |
| rs28567576 | 5 | 136440247 | SPOCK1 | ENSG00000152377.8 | 1.405E-6 | 0.01 | 493821 | gtex_brain_putamen_basal |
| rs66472723 | 5 | 136440304 | SPOCK1 | ENSG00000152377.8 | 3.621E-6 | 0.01 | 493764 | gtex_brain_putamen_basal |
| rs1468293 | 5 | 136441222 | SPOCK1 | ENSG00000152377.8 | 3.575E-6 | 0.01 | 492846 | gtex_brain_putamen_basal |
| rs17522016 | 5 | 136447232 | SPOCK1 | ENSG00000152377.8 | 1.911E-6 | 0.01 | 486836 | gtex_brain_putamen_basal |
| rs68027902 | 5 | 136448540 | SPOCK1 | ENSG00000152377.8 | 2.408E-6 | 0.01 | 485528 | gtex_brain_putamen_basal |
| rs757423 | 5 | 136449299 | SPOCK1 | ENSG00000152377.8 | 2.976E-6 | 0.01 | 484769 | gtex_brain_putamen_basal |
| rs2905968 | 5 | 136457836 | SPOCK1 | ENSG00000152377.8 | 3.267E-6 | 0.01 | 476232 | gtex_brain_putamen_basal |
| rs757424 | 5 | 136459562 | SPOCK1 | ENSG00000152377.8 | 9.726E-7 | 0.01 | 474506 | gtex_brain_putamen_basal |
| rs916980 | 5 | 136460641 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 473427 | gtex_brain_putamen_basal |
| rs2905971 | 5 | 136461551 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 472517 | gtex_brain_putamen_basal |
| rs2905973 | 5 | 136462383 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 471685 | gtex_brain_putamen_basal |
| rs2966729 | 5 | 136462509 | SPOCK1 | ENSG00000152377.8 | 1.762E-6 | 0.01 | 471559 | gtex_brain_putamen_basal |
| rs2966730 | 5 | 136464224 | SPOCK1 | ENSG00000152377.8 | 1.762E-6 | 0.01 | 469844 | gtex_brain_putamen_basal |
| rs2905976 | 5 | 136464933 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 469135 | gtex_brain_putamen_basal |
| rs2905978 | 5 | 136466480 | SPOCK1 | ENSG00000152377.8 | 5.449E-6 | 0.01 | 467588 | gtex_brain_putamen_basal |
| rs2240223 | 5 | 136466581 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 467487 | gtex_brain_putamen_basal |
| rs2240224 | 5 | 136466764 | SPOCK1 | ENSG00000152377.8 | 9.745E-7 | 0.01 | 467304 | gtex_brain_putamen_basal |
| rs6596366 | 5 | 136467795 | SPOCK1 | ENSG00000152377.8 | 1.095E-6 | 0.01 | 466273 | gtex_brain_putamen_basal |
| rs13186295 | 5 | 136468871 | SPOCK1 | ENSG00000152377.8 | 2.66E-6 | 0.01 | 465197 | gtex_brain_putamen_basal |
| rs34812669 | 5 | 136481759 | SPOCK1 | ENSG00000152377.8 | 5.449E-6 | 0.01 | 452309 | gtex_brain_putamen_basal |
| rs1434662 | 5 | 136489133 | SPOCK1 | ENSG00000152377.8 | 1.343E-6 | 0.01 | 444935 | gtex_brain_putamen_basal |
| rs565009222 | 5 | 136496186 | SPOCK1 | ENSG00000152377.8 | 5.712E-6 | 0.01 | 437882 | gtex_brain_putamen_basal |
| rs7713012 | 5 | 136502945 | SPOCK1 | ENSG00000152377.8 | 1.343E-6 | 0.01 | 431123 | gtex_brain_putamen_basal |
| rs6882509 | 5 | 136504046 | SPOCK1 | ENSG00000152377.8 | 4.451E-7 | 0.01 | 430022 | gtex_brain_putamen_basal |
| rs35890961 | 5 | 136505153 | SPOCK1 | ENSG00000152377.8 | 1.343E-6 | 0.01 | 428915 | gtex_brain_putamen_basal |
| rs68178906 | 5 | 136512226 | SPOCK1 | ENSG00000152377.8 | 1.482E-6 | 0.01 | 421842 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
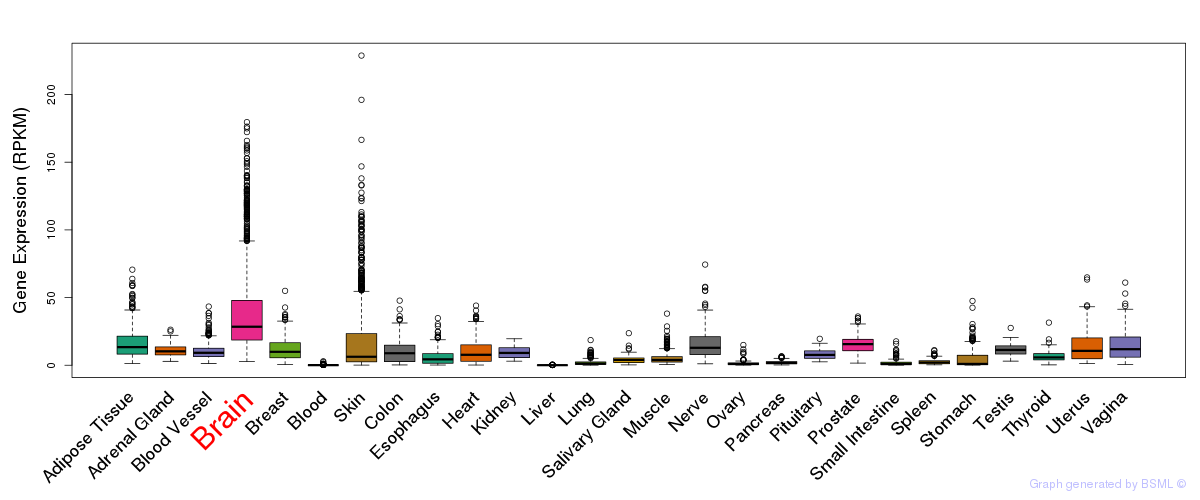
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9181134 |
| GO:0007155 | cell adhesion | TAS | 8389704 | |
| GO:0008283 | cell proliferation | TAS | 8389704 | |
| GO:0006928 | cell motion | TAS | 8389704 | |
| GO:0007275 | multicellular organismal development | TAS | 8389704 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN IMATINIB RESISTANCE UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN GIST MORPHOLOGICAL SWITCH | 15 | 11 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS DN | 47 | 33 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 SIGNALING VIA CTNNB1 | 83 | 58 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE UP | 31 | 21 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 566 | 572 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-135 | 944 | 950 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-139 | 429 | 435 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-23 | 3260 | 3267 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 3260 | 3266 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-326 | 150 | 156 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-375 | 2144 | 2150 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.