Gene Page: BRCA1
Summary ?
| GeneID | 672 |
| Symbol | BRCA1 |
| Synonyms | BRCAI|BRCC1|BROVCA1|FANCS|IRIS|PNCA4|PPP1R53|PSCP|RNF53 |
| Description | breast cancer 1 |
| Reference | MIM:113705|HGNC:HGNC:1100|Ensembl:ENSG00000012048|HPRD:00218|Vega:OTTHUMG00000157426 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.013 |
| Sherlock p-value | 0.658 |
| Fetal beta | 1.161 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1208 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25738236 | 17 | 41278652 | BRCA1;NBR2 | 5.63E-5 | 0.416 | 0.022 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
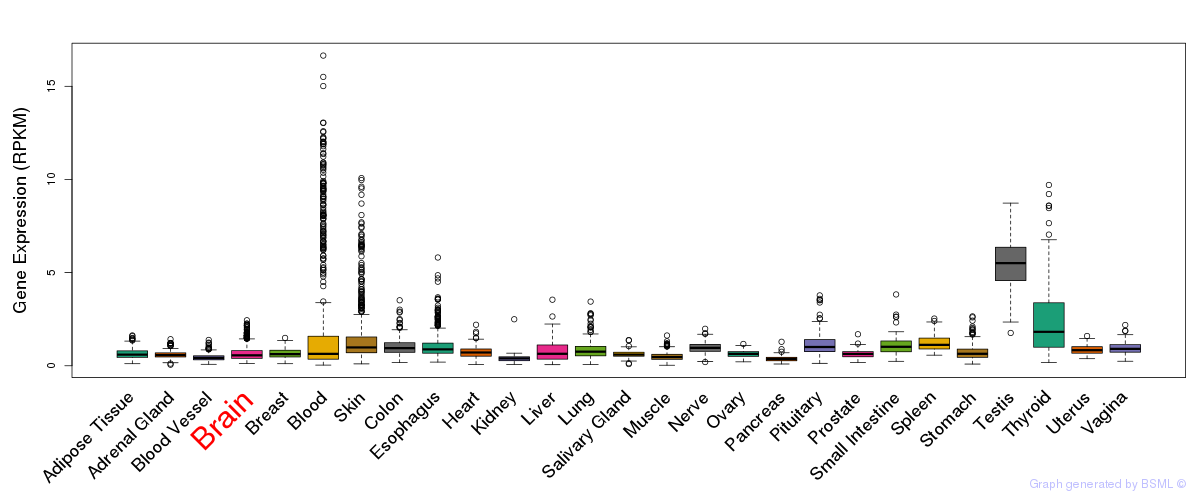
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003684 | damaged DNA binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004842 | ubiquitin-protein ligase activity | NAS | 15905410 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008270 | zinc ion binding | TAS | 8944023 | |
| GO:0015631 | tubulin binding | NAS | 12214252 | |
| GO:0019899 | enzyme binding | IPI | 15965487 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000075 | cell cycle checkpoint | NAS | - | |
| GO:0000724 | double-strand break repair via homologous recombination | IDA | 17349954 | |
| GO:0009048 | dosage compensation, by inactivation of X chromosome | IEA | - | |
| GO:0006359 | regulation of transcription from RNA polymerase III promoter | TAS | 10918303 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 10910365 | |
| GO:0006301 | postreplication repair | IDA | 17349954 | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0006260 | DNA replication | IEA | - | |
| GO:0008630 | DNA damage response, signal transduction resulting in induction of apoptosis | IDA | 14654789 | |
| GO:0007098 | centrosome cycle | IEA | - | |
| GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | TAS | 10918303 | |
| GO:0007059 | chromosome segregation | IMP | 15965487 | |
| GO:0042981 | regulation of apoptosis | TAS | 10918303 | |
| GO:0016481 | negative regulation of transcription | IDA | 16288014 | |
| GO:0042127 | regulation of cell proliferation | TAS | 10918303 | |
| GO:0043009 | chordate embryonic development | IEA | - | |
| GO:0031398 | positive regulation of protein ubiquitination | IDA | 15965487 | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045739 | positive regulation of DNA repair | NAS | 12242698 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | NAS | 15572661 | |
| GO:0045717 | negative regulation of fatty acid biosynthetic process | IMP | 16326698 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0043627 | response to estrogen stimulus | IDA | 8895509 | |
| GO:0046600 | negative regulation of centriole replication | NAS | 12214252 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000151 | ubiquitin ligase complex | NAS | 14976165 | |
| GO:0000793 | condensed chromosome | IEA | - | |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 10918303 | |
| GO:0005654 | nucleoplasm | EXP | 10550055 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0008274 | gamma-tubulin ring complex | NAS | 12214252 | |
| GO:0031436 | BRCA1-BARD1 complex | IDA | 15265711 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Affinity Capture-Western | BioGRID | 12024016 |
| ACACA | ACAC | ACC | ACC1 | ACCA | acetyl-Coenzyme A carboxylase alpha | - | HPRD,BioGRID | 12360400 |
| ACACA | ACAC | ACC | ACC1 | ACCA | acetyl-Coenzyme A carboxylase alpha | BRCA1 interacts with acetyl-CoA carboxylase. | BIND | 12360400 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Reconstituted Complex | BioGRID | 10542266 |
| APLP2 | APPH | APPL2 | CDEBP | amyloid beta (A4) precursor-like protein 2 | Far Western | BioGRID | 11746496 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11016951 |11085509 |
| ATF1 | EWS-ATF1 | FUS/ATF-1 | TREB36 | activating transcription factor 1 | - | HPRD,BioGRID | 10945975 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Affinity Capture-Western Co-purification Protein-peptide Reconstituted Complex | BioGRID | 10550055 |10608806 |10783165 |10866324 |11016625 |11114888 |11278964 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | - | HPRD | 2001833 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | BRCA1 physically and functionally interacts with ATR during genotoxic stress | BIND | 11016625 |11278964 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Affinity Capture-Western Protein-peptide Reconstituted Complex | BioGRID | 10608806 |11016625 |11114888 |11278964 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | Affinity Capture-Western Reconstituted Complex | BioGRID | 14990569 |
| BACH1 | - | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | - | HPRD,BioGRID | 11301010 |
| BAP1 | DKFZp686N04275 | FLJ35406 | FLJ37180 | HUCEP-13 | KIAA0272 | UCHL2 | hucep-6 | BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) | - | HPRD | 9528852 |
| BARD1 | - | BRCA1 associated RING domain 1 | Affinity Capture-MS Affinity Capture-Western Co-crystal Structure Co-localization Co-purification FRET Reconstituted Complex Two-hybrid | BioGRID | 8944023 |9342365 |9738006 |9788437 |10026184 |10477523 |10635334 |11257228 |11278247 |11301010 |11498787 |11504724 |11526114 |11573085 |11773071 |11925436 |11927591 |12431996 |12438698 |12485996 |12700228 |12732733 |12887909 |12890688 |12951069 |14636569 |14638690 |14647430 |15159397 |15184379 |
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD | 8944023 |10026184 |11573085 |11773071 |12431996 |
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD | 8944023 |10026184 |11773071 |12431996 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | Co-purification | BioGRID | 10783165 |
| BRAP | BRAP2 | IMP | RNF52 | BRCA1 associated protein | - | HPRD,BioGRID | 9497340 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Co-crystal Structure Protein-peptide Reconstituted Complex | BioGRID | 9525870 |11573086 |14578343 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 possesses E3 ubiquitin ligase activity and auto-ubiquitinates | BIND | 11320250 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | - | HPRD,BioGRID | 9774970 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | BRCA1 interacts with BRCA2. | BIND | 9774970 |
| BRCC3 | BRCC36 | C6.1A | CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BRE | BRCC4 | BRCC45 | brain and reproductive organ-expressed (TNFRSF1A modulator) | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BRIP1 | BACH1 | FANCJ | FLJ90232 | MGC126521 | MGC126523 | OF | BRCA1 interacting protein C-terminal helicase 1 | Affinity Capture-MS Affinity Capture-Western Co-crystal Structure Protein-peptide Reconstituted Complex | BioGRID | 11877378 |14576433 |14578343 |15133502 |15208681 |15242590 |
| BRIP1 | BACH1 | FANCJ | FLJ90232 | MGC126521 | MGC126523 | OF | BRCA1 interacting protein C-terminal helicase 1 | - | HPRD | 11301010 |
| CCNA1 | - | cyclin A1 | BRCA1 associates with Cyclin A | BIND | 9244350 |10373534 |
| CCNA1 | - | cyclin A1 | Reconstituted Complex | BioGRID | 9244350 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD,BioGRID | 9244350 |10373534 |
| CCNB1 | CCNB | cyclin B1 | - | HPRD,BioGRID | 9244350 |
| CCNB1 | CCNB | cyclin B1 | BRCA1 is a tyrosine phosphoprotein that associates with Cyclin B1 | BIND | 9244350 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 9244350 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | BRCA1 associates with Cyclin D1 | BIND | 9244350 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 17525332 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 9244350 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | BRCA1 associates with CDK2 | BIND | 9244350 |10373534 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8764100 |9244350 |10373534 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD,BioGRID | 9244350 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | BRCA1 associates with CDK4 | BIND | 9244350 |
| CDK5 | PSSALRE | cyclin-dependent kinase 5 | Reconstituted Complex | BioGRID | 9244350 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Co-purification | BioGRID | 9159119 |
| CDS1 | CDS | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 | Reconstituted Complex | BioGRID | 12438214 |
| CHEK2 | CDS1 | CHK2 | HuCds1 | LFS2 | PP1425 | RAD53 | CHK2 checkpoint homolog (S. pombe) | - | HPRD,BioGRID | 10724175 |
| CLSPN | CLASPIN | MGC131612 | MGC131613 | MGC131615 | claspin homolog (Xenopus laevis) | - | HPRD,BioGRID | 15096610 |
| COBRA1 | DKFZp586B0519 | KIAA1182 | NELF-B | NELFB | cofactor of BRCA1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11739404 |
| COBRA1 | DKFZp586B0519 | KIAA1182 | NELF-B | NELFB | cofactor of BRCA1 | - | HPRD | 12612062 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Phenotypic Enhancement | BioGRID | 10945975 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 10655477 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with BRCA1. This interaction was modelled on a demonstrated interaction between human BRCA1 and mouse CBP. | BIND | 10655477 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 10403822 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | Reconstituted Complex Two-hybrid | BioGRID | 10403822 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | BRCA1 is phosphorylated by Casein kinase 2 through a physical interaction with CSNK2B | BIND | 10403822 |
| CSTF1 | CstF-50 | CstFp50 | cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa | Reconstituted Complex | BioGRID | 11257228 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Affinity Capture-Western Co-purification | BioGRID | 10477523 |11257228 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | - | HPRD,BioGRID | 10196224 |
| CTCFL | BORIS | CTCF-T | MGC163358 | dJ579F20.2 | CCCTC-binding factor (zinc finger protein)-like | Protein-peptide | BioGRID | 14578343 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD | 9662397|12592385 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9662397 |12592385 |
| DNAJA3 | FLJ45758 | TID1 | hTid-1 | DnaJ (Hsp40) homolog, subfamily A, member 3 | Protein-peptide | BioGRID | 14578343 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | BRCA1 associates with E2F-1 | BIND | 9244350 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | - | HPRD | 9244350 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | BRCA1 associates with E2F-4 | BIND | 9244350 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | - | HPRD,BioGRID | 9244350 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD | 11313879 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11313879 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 10655477 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with BRCA1. | BIND | 10655477 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 10334989 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 11244506 |11493692 |11782371 |12400015 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | Reconstituted Complex | BioGRID | 11313879 |
| ETS2 | ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | ETS2 (Ets-2) interacts with BRCA1 promoter. This interaction was modeled on a demonstrated interaction between ETS2 from an unspecified species and BRCA1 promoter from an unspecified species. | BIND | 12637547 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | BRCA1 interacts with FANCA. | BIND | 12354784 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | - | HPRD,BioGRID | 12354784 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Co-localization Reconstituted Complex Two-hybrid | BioGRID | 12887909 |14499622 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | - | HPRD | 11239454 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14550570 |14986435 |
| FLI1 | EWSR2 | SIC-1 | Friend leukemia virus integration 1 | Reconstituted Complex | BioGRID | 11313879 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 11602572 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | Co-purification | BioGRID | 9159119 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Co-purification | BioGRID | 9159119 |
| GTF2H4 | TFB2 | TFIIH | general transcription factor IIH, polypeptide 4, 52kDa | Co-purification | BioGRID | 9159119 |
| H2AFX | H2A.X | H2A/X | H2AX | H2A histone family, member X | Co-localization Reconstituted Complex | BioGRID | 10959836 |11927591 |12485996 |
| H2AFY | H2A.y | H2A/y | H2AF12M | H2AFJ | MACROH2A1.1 | mH2A1 | macroH2A1.2 | H2A histone family, member Y | Co-localization | BioGRID | 12419249 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | BRCA1 interacts with HDAC1 | BIND | 10220405 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 10220405 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 10220405 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | BRCA1 interacts with HDAC2 | BIND | 10220405 |
| HIST2H2AC | H2A | H2A-GL101 | H2A/q | H2AFQ | MGC74460 | histone cluster 2, H2ac | Reconstituted Complex | BioGRID | 11927591 |
| HIST2H3C | H3 | H3.2 | H3/M | H3F2 | H3FM | MGC9629 | histone cluster 2, H3c | Co-localization | BioGRID | 12419249 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Two-hybrid | BioGRID | 9738006 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 11163768 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 11163768 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | BRCA1 interacts with c-Jun. | BIND | 12080089 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 12080089 |
| JUNB | AP-1 | jun B proto-oncogene | BRCA1 interacts with JunB. | BIND | 12080089 |
| JUNB | AP-1 | jun B proto-oncogene | - | HPRD,BioGRID | 12080089 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD,BioGRID | 12080089 |
| JUND | AP-1 | jun D proto-oncogene | BRCA1 interacts with JunD. | BIND | 12080089 |
| KIF1B | CMT2 | CMT2A | CMT2A1 | FLJ23699 | HMSNII | KIAA0591 | KIAA1448 | KLP | MGC134844 | kinesin family member 1B | Protein-peptide | BioGRID | 14578343 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | Two-hybrid | BioGRID | 8955125 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | - | HPRD | 9497340 |
| LMO4 | - | LIM domain only 4 | - | HPRD,BioGRID | 11751867 |
| LMO4 | - | LIM domain only 4 | The LIM domain protein LMO4 interacts with BRCA1 | BIND | 11751867 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | Affinity Capture-Western Two-hybrid | BioGRID | 15205325 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | BRCA1 interacts with MEKK3. | BIND | 15205325 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | Protein-peptide | BioGRID | 14578343 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | - | HPRD | 12607005 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | Affinity Capture-Western | BioGRID | 17525332 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | BRCA1 interacts with TRAP220. | BIND | 15208681 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 15208681 |
| MED13 | ARC250 | DRIP250 | HSPC221 | KIAA0593 | THRAP1 | TRAP240 | mediator complex subunit 13 | Affinity Capture-MS | BioGRID | 15208681 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | Affinity Capture-MS Co-purification | BioGRID | 11504724 |12154023 |15208681 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | - | HPRD,BioGRID | 9159119 |
| MED23 | CRSP130 | CRSP133 | CRSP3 | DKFZp434H0117 | DRIP130 | SUR2 | mediator complex subunit 23 | Affinity Capture-MS | BioGRID | 15208681 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15208681 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | - | HPRD,BioGRID | 10783165 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Affinity Capture-Western Co-localization Co-purification Reconstituted Complex | BioGRID | 10426999 |10783165 |11353843 |11504724 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | - | HPRD,BioGRID | 11498787 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | BRCA1 interacts with MSH2 in the absence of ADP-MSH2 interaction. | BIND | 10783165 |
| MSH3 | DUP | MGC163306 | MGC163308 | MRP1 | mutS homolog 3 (E. coli) | Protein-peptide Reconstituted Complex | BioGRID | 11498787 |14578343 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | BRCA1 associates with MSH6. | BIND | 10783165 |11498787 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | - | HPRD,BioGRID | 11498787 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 9788437 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | BRCA1 association with c-Myc, a proto-oncogene that is implicated in tumorigenesis, embryonic development and apoptosis, inhibits its transcriptional and transforming activity in cells. | BIND | 9788437 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | - | HPRD,BioGRID | 10426999 |10783165 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | Protein-peptide Reconstituted Complex | BioGRID | 11085509 |14578343 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | Protein-peptide | BioGRID | 14578343 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 12700228 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | - | HPRD,BioGRID | 11777930 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | BRCA1 interacts with NF-YA | BIND | 11777930 |
| NMI | - | N-myc (and STAT) interactor | - | HPRD,BioGRID | 11916966 |
| NMI | - | N-myc (and STAT) interactor | BRCA1 interacts with Nmi. | BIND | 11916966 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 15184379 |
| NUFIP1 | NUFIP | bA540M5.1 | nuclear fragile X mental retardation protein interacting protein 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15107825 |
| NUFIP1 | NUFIP | bA540M5.1 | nuclear fragile X mental retardation protein interacting protein 1 | BRCA1 interacts with NUFIP. | BIND | 15107825 |
| NUP153 | HNUP153 | N153 | nucleoporin 153kDa | Protein-peptide | BioGRID | 14578343 |
| ORC2L | ORC2 | origin recognition complex, subunit 2-like (yeast) | Affinity Capture-Western | BioGRID | 17525332 |
| ORC3L | LAT | LATHEO | ORC3 | origin recognition complex, subunit 3-like (yeast) | Affinity Capture-Western | BioGRID | 17525332 |
| PCTK1 | FLJ16665 | PCTAIRE | PCTAIRE1 | PCTGAIRE | PCTAIRE protein kinase 1 | Two-hybrid | BioGRID | 9738006 |
| PEG3 | DKFZp781A095 | KIAA0287 | PW1 | ZSCAN24 | paternally expressed 3 | Far Western | BioGRID | 11746496 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 4506230 |9159119 |12154023|10725406 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 4506230 |9159119 |12154023 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western Co-purification | BioGRID | 9159119 |11504724 |12955082 |14506230 |
| POLR2K | ABC10-alpha | RPABC4 | RPB10alpha | RPB12 | RPB7.0 | hRPB7.0 | hsRPB10a | polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa | - | HPRD | 10725406 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Affinity Capture-Western | BioGRID | 11777930 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | BRCA1 interacts with Oct-1. | BIND | 11777930 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | Affinity Capture-Western Reconstituted Complex | BioGRID | 12438214 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | Protein-peptide | BioGRID | 10608806 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10426999 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 9774970 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | BRCA1 and Rad51 associate in mitotic and meiotic cells | BIND | 9008167 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD | 9008167 |9774970|9774970 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 10220405 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | BRCA1 interacts with Rb. | BIND | 10518542 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | BRCA1 interacts with RbAp48. | BIND | 10220405 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | - | HPRD,BioGRID | 10220405 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | BRCA1 interacts with RbAp46. | BIND | 10220405 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | - | HPRD,BioGRID | 10220405 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | - | HPRD | 9738006 |10196224 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | Affinity Capture-Western Protein-peptide Reconstituted Complex Two-hybrid | BioGRID | 9738006 |9811458 |10196224 |10764811 |10910365 |11689934 |14578343 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | BRCA1 interacts with CtIP. | BIND | 9811458 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | Co-localization Reconstituted Complex | BioGRID | 11521194 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | Co-localization Reconstituted Complex | BioGRID | 11521194 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 12700228 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | - | HPRD,BioGRID | 10783165 |
| RPL31 | MGC88191 | ribosomal protein L31 | Far Western Reconstituted Complex | BioGRID | 11746496 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | BRCA1 interacts with Smad2. This interaction was modeled on a demonstrated interaction between BRCA1 from human and Smad2 from an unspecified species. | BIND | 15735739 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | BRCA1 interacts with Smad3. | BIND | 15735739 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | BRCA1 interacts with Smad4. This interaction was modeled on a demonstrated interaction between BRCA1 from human and Smad4 from an unspecified species. | BIND | 15735739 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Affinity Capture-Western | BioGRID | 15034933 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 10943845 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Co-purification | BioGRID | 10943845 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Co-purification | BioGRID | 10943845 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Co-purification | BioGRID | 10943845 |
| SMARCD2 | BAF60B | CRACD2 | PRO2451 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | Co-purification | BioGRID | 10943845 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | - | HPRD | 11877377 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 10720440 |
| SP1 | - | Sp1 transcription factor | BRCA1 interacts with Sp1. | BIND | 12706836 |
| SP1 | - | Sp1 transcription factor | Affinity Capture-Western | BioGRID | 12706836 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 10792030 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD | 11163768 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 12459499 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | TOP2A (Topo II) interacts with BRCA1 | BIND | 15965487 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | BRCA1a interacts with p53. | BIND | 9926942 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | BRCA1 physically associates with p53 and stimulates its transcriptional activity | BIND | 9582019 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9482880 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western | BioGRID | 17525332 |
| TUBG1 | TUBG | TUBGCP1 | tubulin, gamma 1 | - | HPRD,BioGRID | 9789027 |11691781 |
| UBA1 | A1S9 | A1S9T | A1ST | AMCX1 | GXP1 | MGC4781 | SMAX2 | UBA1A | UBE1 | UBE1X | ubiquitin-like modifier activating enzyme 1 | Reconstituted Complex | BioGRID | 11278247 |11927591 |12438698 |12485996 |12887909 |14638690 |15184379 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | BRCA1 interacts with Ub. This interaction was modelled on a demonstrated interaction between human BRCA1 and Ub from an unspecified source. | BIND | 11320250 |
| UBE2D1 | E2(17)KB1 | SFT | UBC4/5 | UBCH5 | UBCH5A | ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) | Reconstituted Complex | BioGRID | 11278247 |11927591 |12438698 |12485996 |12732733 |12887909 |12890688 |14636569 |14638690 |15184379 |
| UBE2D3 | E2(17)KB3 | MGC43926 | MGC5416 | UBC4/5 | UBCH5C | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | - | HPRD | 12732733 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD,BioGRID | 12732733 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | Reconstituted Complex | BioGRID | 12890688 |
| USF2 | FIP | bHLHb12 | upstream transcription factor 2, c-fos interacting | - | HPRD,BioGRID | 14502648 |
| VCP | IBMPFD | MGC131997 | MGC148092 | MGC8560 | TERA | p97 | valosin-containing protein | - | HPRD,BioGRID | 10855792 |
| WNT2B | WNT13 | XWNT2 | wingless-type MMTV integration site family, member 2B | Far Western | BioGRID | 11746496 |
| XIST | DKFZp779P0129 | DXS1089 | DXS399E | NCRNA00001 | SXI1 | XCE | XIC | swd66 | X (inactive)-specific transcript (non-protein coding) | Co-localization | BioGRID | 12419249 |15065664 |
| XRCC1 | RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 | - | HPRD | 11352725 |
| ZCCHC8 | DKFZp434E2220 | zinc finger, CCHC domain containing 8 | Affinity Capture-Western | BioGRID | 17525332 |
| ZNF350 | ZBRK1 | ZFQR | zinc finger protein 350 | - | HPRD,BioGRID | 11090615 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID AURORA A PATHWAY | 31 | 20 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME FANCONI ANEMIA PATHWAY | 25 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| BUSA SAM68 TARGETS DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| KONG E2F3 TARGETS | 97 | 58 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 12 | 11 | 8 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 6HR | 27 | 18 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 48HR | 9 | 6 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE UP | 34 | 17 | All SZGR 2.0 genes in this pathway |
| KARAKAS TGFB1 SIGNALING | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| OHM EMBRYONIC CARCINOMA UP | 6 | 5 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS DN | 21 | 15 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS UP | 29 | 23 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA MAST CELL | 46 | 34 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| UROSEVIC RESPONSE TO IMIQUIMOD | 23 | 14 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE LITERATURE | 44 | 25 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| YU BAP1 TARGETS | 29 | 21 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-132/212 | 1242 | 1248 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.