Gene Page: STAU1
Summary ?
| GeneID | 6780 |
| Symbol | STAU1 |
| Synonyms | PPP1R150|STAU |
| Description | staufen double-stranded RNA binding protein 1 |
| Reference | MIM:601716|HGNC:HGNC:11370|Ensembl:ENSG00000124214|HPRD:03422|Vega:OTTHUMG00000032691 |
| Gene type | protein-coding |
| Map location | 20q13.1 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.963 |
| Fetal beta | 0.639 |
| eGene | Frontal Cortex BA9 |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0179 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
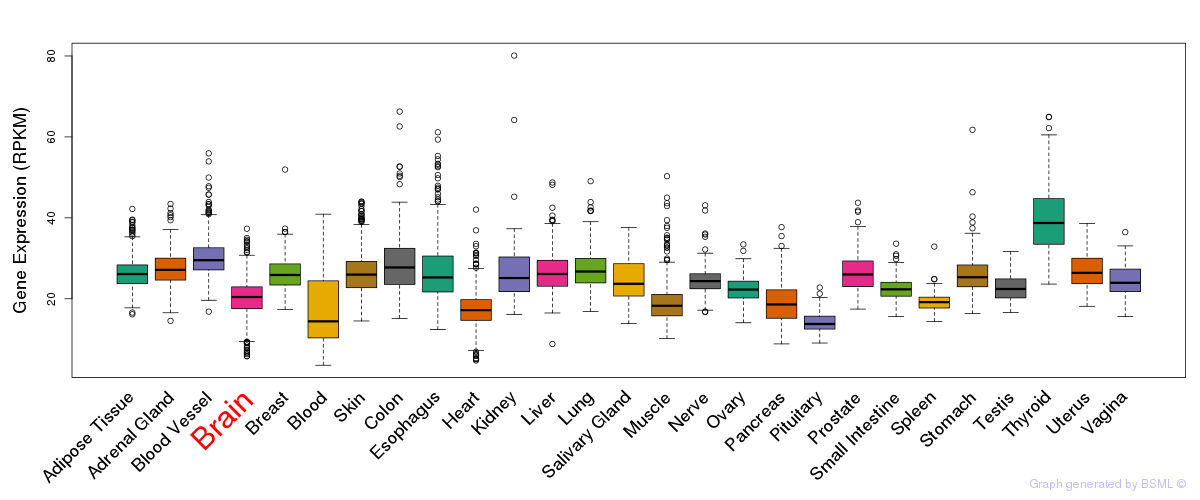
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LGALS1 | 0.53 | 0.57 |
| AGPAT2 | 0.51 | 0.53 |
| MRPL41 | 0.50 | 0.53 |
| MSRB2 | 0.50 | 0.57 |
| PVALB | 0.49 | 0.55 |
| CRYBA2 | 0.48 | 0.53 |
| SDF2L1 | 0.48 | 0.42 |
| SMPX | 0.47 | 0.43 |
| SNCG | 0.46 | 0.52 |
| DAPL1 | 0.46 | 0.37 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AOF1 | -0.42 | -0.57 |
| THOC2 | -0.42 | -0.59 |
| MAP4K4 | -0.42 | -0.55 |
| PTPN1 | -0.42 | -0.56 |
| GNB4 | -0.42 | -0.57 |
| PHF3 | -0.41 | -0.57 |
| LATS1 | -0.41 | -0.57 |
| TOP1 | -0.41 | -0.58 |
| RSF1 | -0.41 | -0.56 |
| HP1BP3 | -0.41 | -0.55 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003725 | double-stranded RNA binding | IEA | - | |
| GO:0003725 | double-stranded RNA binding | TAS | 10022909 | |
| GO:0005515 | protein binding | IPI | 15303970 |15680326 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008298 | intracellular mRNA localization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005791 | rough endoplasmic reticulum | TAS | 10022909 | |
| GO:0005875 | microtubule associated complex | TAS | 10022909 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0030529 | ribonucleoprotein complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR2 TARGETS UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-3p | 1039 | 1046 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA | ||||
| miR-142-5p | 1498 | 1505 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-326 | 458 | 464 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-330 | 60 | 67 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-361 | 80 | 86 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-381 | 859 | 865 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-486 | 1043 | 1049 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-495 | 1360 | 1366 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.