Gene Page: STXBP3
Summary ?
| GeneID | 6814 |
| Symbol | STXBP3 |
| Synonyms | MUNC18-3|MUNC18C|PSP|UNC-18C |
| Description | syntaxin binding protein 3 |
| Reference | MIM:608339|HGNC:HGNC:11446|Ensembl:ENSG00000116266|HPRD:10517|Vega:OTTHUMG00000011122 |
| Gene type | protein-coding |
| Map location | 1p13.3 |
| Pascal p-value | 0.298 |
| Sherlock p-value | 0.517 |
| Fetal beta | -1.128 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
| Support | CANABINOID DOPAMINE EXOCYTOSIS METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06527153 | 1 | 109289113 | STXBP3 | 5.21E-8 | -0.007 | 1.37E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6599250 | chr3 | 38784028 | STXBP3 | 6814 | 0.17 | trans |
Section II. Transcriptome annotation
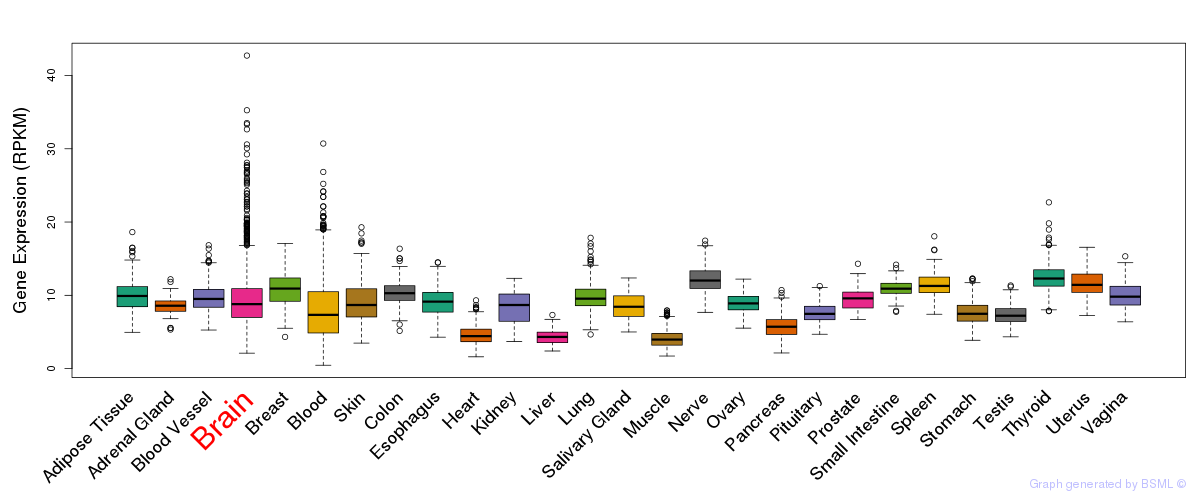
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM130 | 0.93 | 0.96 |
| MFSD4 | 0.91 | 0.86 |
| SYN1 | 0.90 | 0.95 |
| SV2B | 0.90 | 0.93 |
| AK5 | 0.89 | 0.85 |
| MAP2K1 | 0.89 | 0.93 |
| EPDR1 | 0.89 | 0.89 |
| SERINC3 | 0.89 | 0.92 |
| YWHAH | 0.89 | 0.91 |
| AGBL4 | 0.89 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.51 | -0.57 |
| C9orf46 | -0.46 | -0.42 |
| FAM36A | -0.44 | -0.32 |
| GTF3C6 | -0.44 | -0.39 |
| C21orf57 | -0.43 | -0.40 |
| RBMX2 | -0.43 | -0.43 |
| RPL23A | -0.43 | -0.44 |
| RPS19P3 | -0.42 | -0.47 |
| SH3BP2 | -0.42 | -0.41 |
| DYNLT1 | -0.42 | -0.45 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | - | HPRD | 12065586 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | - | HPRD | 12065586 |
| GEMIN5 | DKFZp586M1824 | MGC142174 | gem (nuclear organelle) associated protein 5 | - | HPRD | 12065586 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD | 10194441 |
| PRKCSH | AGE-R2 | G19P1 | PCLD | PLD1 | protein kinase C substrate 80K-H | PRKCSH (80K-H) interacts with STXBP3 (Munc18c).This interaction was modelled on a demonstrated interaction between human 80K-H and hamster Munc18c. | BIND | 15707389 |
| SIP1 | GEMIN2 | SIP1-delta | survival of motor neuron protein interacting protein 1 | - | HPRD | 12065586 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD | 12065586 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | Affinity Capture-Western | BioGRID | 12773094 |
| SNRPB | COD | SNRPB1 | SmB/SmB' | snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 | - | HPRD | 12065586 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | - | HPRD | 12065586 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | - | HPRD | 12065586 |
| SNTG1 | G1SYN | SYN4 | syntrophin, gamma 1 | Reconstituted Complex | BioGRID | 12832401 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | Affinity Capture-Western | BioGRID | 12773094 |
| STX4 | STX4A | p35-2 | syntaxin 4 | - | HPRD | 10194441 |
| STX4 | STX4A | p35-2 | syntaxin 4 | Affinity Capture-Western Two-hybrid | BioGRID | 12773094 |12832401 |
| STXBP2 | Hunc18b | MUNC18-2 | UNC18-2 | UNC18B | pp10122 | syntaxin binding protein 2 | - | HPRD | 12065586 |
| VAMP2 | FLJ11460 | SYB2 | VAMP-2 | vesicle-associated membrane protein 2 (synaptobrevin 2) | Reconstituted Complex | BioGRID | 12832401 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D1 | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |