Gene Page: MED22
Summary ?
| GeneID | 6837 |
| Symbol | MED22 |
| Synonyms | MED24|SURF5|surf-5 |
| Description | mediator complex subunit 22 |
| Reference | MIM:185641|HGNC:HGNC:11477|Ensembl:ENSG00000148297|HPRD:01713|Vega:OTTHUMG00000020869 |
| Gene type | protein-coding |
| Map location | 9q34.2 |
| Pascal p-value | 0.308 |
| Sherlock p-value | 0.008 |
| Fetal beta | 0.717 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
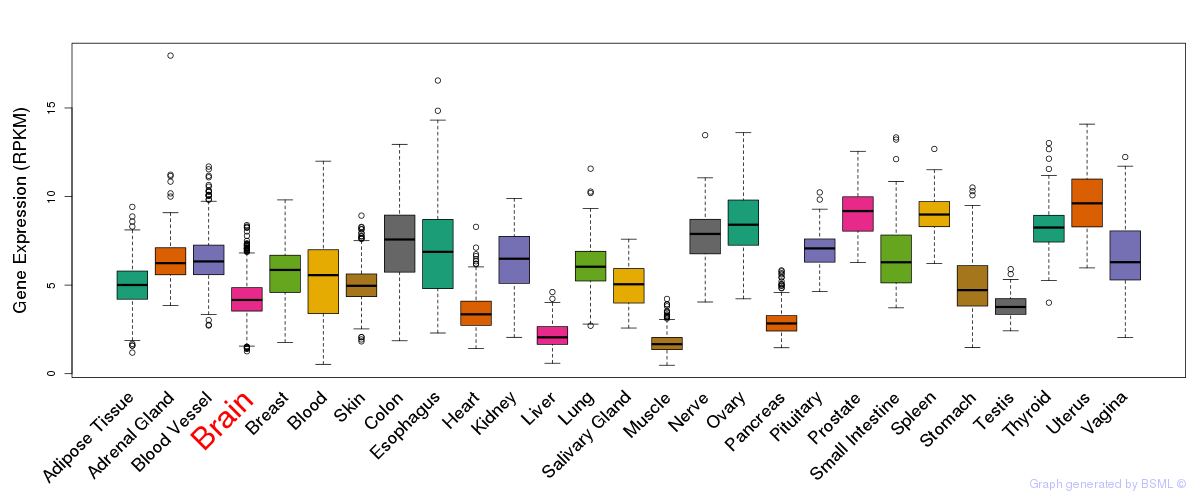
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NONO | 0.96 | 0.93 |
| ANP32A | 0.95 | 0.92 |
| KDM1 | 0.95 | 0.94 |
| INTS7 | 0.95 | 0.94 |
| FUBP1 | 0.95 | 0.95 |
| XRN2 | 0.95 | 0.93 |
| EIF4B | 0.94 | 0.93 |
| DHX9 | 0.94 | 0.95 |
| MSH6 | 0.94 | 0.94 |
| RBMX | 0.94 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.70 | -0.81 |
| AF347015.31 | -0.70 | -0.91 |
| FXYD1 | -0.70 | -0.92 |
| AIFM3 | -0.70 | -0.81 |
| TSC22D4 | -0.70 | -0.84 |
| MT-CO2 | -0.69 | -0.91 |
| AF347015.33 | -0.69 | -0.89 |
| C5orf53 | -0.69 | -0.76 |
| PTH1R | -0.69 | -0.79 |
| IFI27 | -0.69 | -0.90 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| GOLM1 | C9orf155 | FLJ22634 | FLJ23608 | GOLPH2 | GP73 | PSEC0257 | bA379P1.3 | golgi membrane protein 1 | Two-hybrid | BioGRID | 16169070 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS | BioGRID | 12584197 |15175163 |
| MED11 | HSPC296 | MGC88387 | mediator complex subunit 11 | - | HPRD,BioGRID | 12584197 |
| MED11 | HSPC296 | MGC88387 | mediator complex subunit 11 | Surf5 interacts with HSPC296. This interaction was modelled on a demonstrated interaction between human Surf5 and mouse HSPC296. | BIND | 12584197 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | TRAP80 interacts with Surf5. | BIND | 12584197 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED25 | ACID1 | ARC92 | DKFZp434K0512 | MGC70671 | P78 | PTOV2 | TCBAP0758 | mediator complex subunit 25 | Affinity Capture-Western | BioGRID | 14638676 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED27 | CRAP34 | CRSP34 | CRSP8 | MGC11274 | TRAP37 | mediator complex subunit 27 | Reconstituted Complex | BioGRID | 12584197 |
| MED27 | CRAP34 | CRSP34 | CRSP8 | MGC11274 | TRAP37 | mediator complex subunit 27 | TRAP37 interacts with Surf5. | BIND | 12584197 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD | 14576168 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| MED30 | MGC9890 | THRAP6 | TRAP25 | mediator complex subunit 30 | - | HPRD,BioGRID | 12584197 |
| MED30 | MGC9890 | THRAP6 | TRAP25 | mediator complex subunit 30 | Surf5 interacts with TRAP25. This interaction was modelled on a demonstrated interaction between human Surf5 and mouse TRAP25. | BIND | 12584197 |
| MED7 | CRSP33 | CRSP9 | MGC12284 | mediator complex subunit 7 | Reconstituted Complex | BioGRID | 12584197 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| THOC7 | FLJ23445 | NIF3L1BP1 | fSAP24 | THO complex 7 homolog (Drosophila) | Reconstituted Complex | BioGRID | 12584197 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4 | 61 | 34 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |