| KEGG WNT SIGNALING PATHWAY
| 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION
| 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS
| 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER
| 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER
| 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER
| 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER
| 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER
| 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA
| 55 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA
| 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC
| 76 | 59 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY
| 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY
| 80 | 60 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP
| 59 | 38 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP
| 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP
| 64 | 37 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP
| 485 | 293 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP
| 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP
| 368 | 234 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP
| 404 | 246 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP
| 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP
| 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP
| 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN
| 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN
| 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP
| 57 | 33 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN
| 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP
| 149 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN
| 770 | 415 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP
| 121 | 72 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL
| 330 | 217 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN
| 514 | 319 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN
| 50 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP
| 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN
| 56 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN
| 298 | 200 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP
| 175 | 116 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C1
| 19 | 15 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1
| 58 | 43 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN
| 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN
| 128 | 93 | All SZGR 2.0 genes in this pathway |
| CHENG RESPONSE TO NICKEL ACETATE
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN
| 160 | 101 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS
| 82 | 55 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV
| 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN
| 195 | 135 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP
| 181 | 112 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN
| 80 | 66 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C
| 463 | 262 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN
| 210 | 128 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP
| 79 | 40 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN
| 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN
| 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP
| 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP
| 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN
| 442 | 275 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP
| 47 | 38 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP
| 93 | 62 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN
| 88 | 61 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN
| 918 | 550 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES
| 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION
| 37 | 20 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN
| 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN
| 244 | 157 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES
| 462 | 273 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS
| 239 | 156 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL
| 160 | 106 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN
| 146 | 88 | All SZGR 2.0 genes in this pathway |
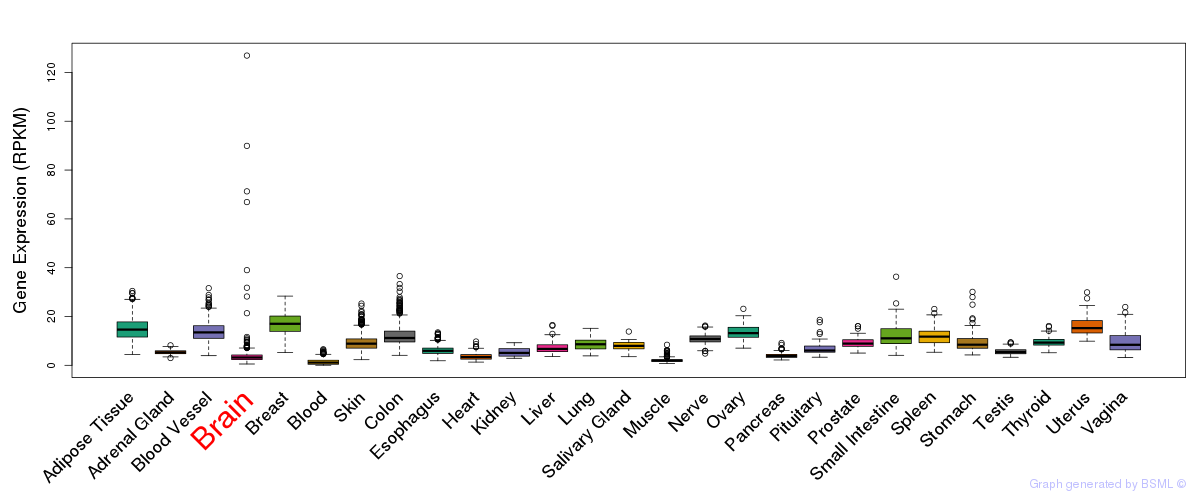
 Differentially methylated gene
Differentially methylated gene