Gene Page: TEAD1
Summary ?
| GeneID | 7003 |
| Symbol | TEAD1 |
| Synonyms | AA|NTEF-1|REF1|TCF-13|TCF13|TEAD-1|TEF-1 |
| Description | TEA domain transcription factor 1 |
| Reference | MIM:189967|HGNC:HGNC:11714|Ensembl:ENSG00000187079|HPRD:01802| |
| Gene type | protein-coding |
| Map location | 11p15.2 |
| Pascal p-value | 0.053 |
| Fetal beta | 0.674 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06829681 | 11 | 12867453 | TEAD1 | 5.3E-5 | -0.565 | 0.022 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
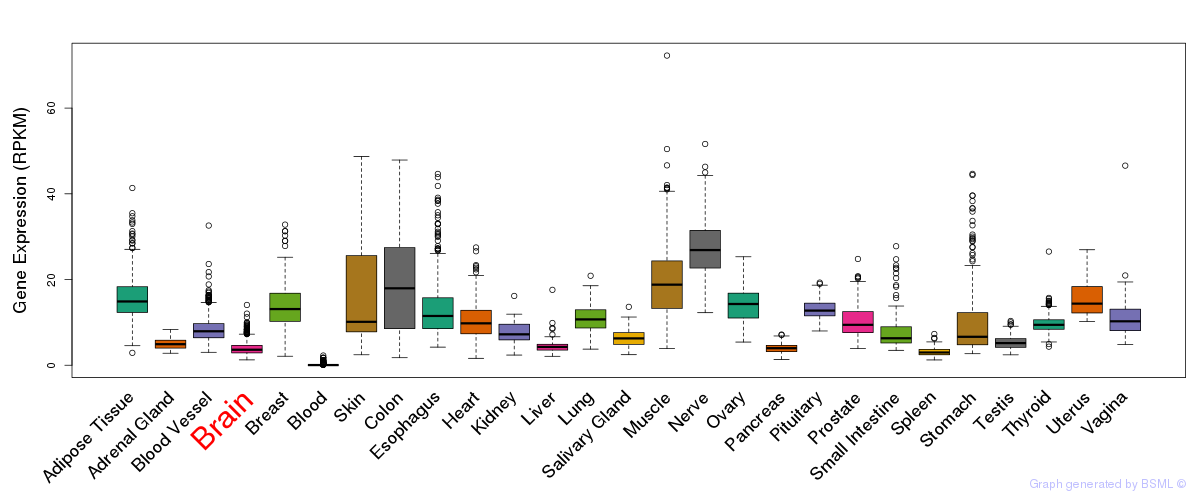
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SFRP4 | 0.69 | 0.62 |
| ECM2 | 0.66 | 0.73 |
| ABCA4 | 0.63 | 0.49 |
| COL8A2 | 0.62 | 0.53 |
| SLC5A5 | 0.61 | 0.59 |
| SLC13A4 | 0.60 | 0.55 |
| ANGPT1 | 0.60 | 0.56 |
| OLFML1 | 0.60 | 0.61 |
| PLBD1 | 0.59 | 0.52 |
| SIGLEC1 | 0.59 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC132872.1 | -0.27 | -0.38 |
| SH2B2 | -0.25 | -0.23 |
| AC005393.1 | -0.25 | -0.21 |
| RPL27 | -0.24 | -0.38 |
| TUBB2B | -0.24 | -0.25 |
| SNHG12 | -0.24 | -0.34 |
| RPL37 | -0.24 | -0.40 |
| RPS19P3 | -0.24 | -0.35 |
| RPL23A | -0.24 | -0.39 |
| CARHSP1 | -0.24 | -0.31 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |