Gene Page: TEC
Summary ?
| GeneID | 7006 |
| Symbol | TEC |
| Synonyms | PSCTK4 |
| Description | tec protein tyrosine kinase |
| Reference | MIM:600583|HGNC:HGNC:11719|Ensembl:ENSG00000135605|HPRD:02786|Vega:OTTHUMG00000128623 |
| Gene type | protein-coding |
| Map location | 4p12 |
| Pascal p-value | 0.217 |
| Fetal beta | -0.048 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
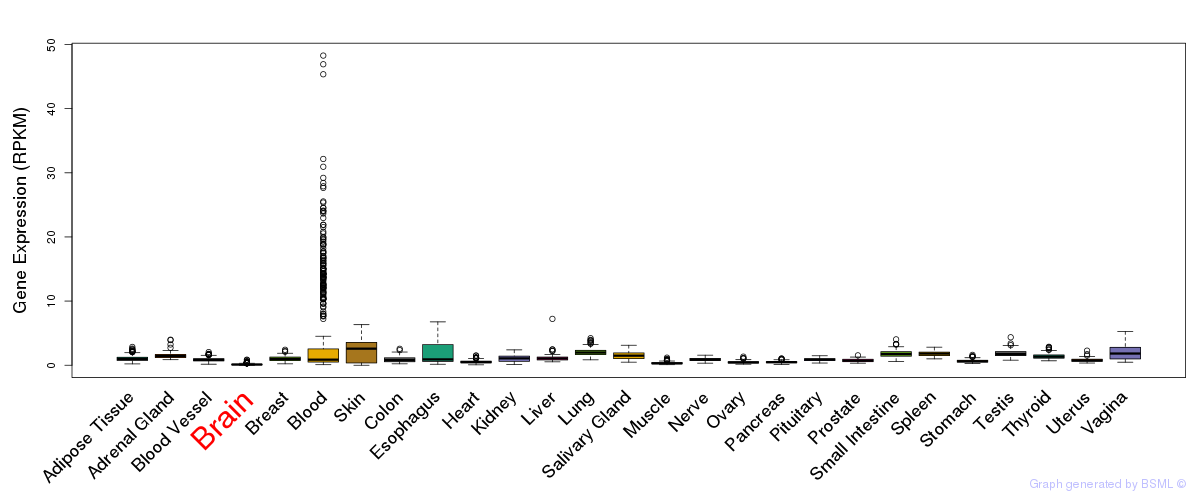
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPRB | 0.84 | 0.88 |
| COBLL1 | 0.82 | 0.88 |
| GBP4 | 0.80 | 0.84 |
| ABCB1 | 0.80 | 0.82 |
| GPR116 | 0.80 | 0.86 |
| FLT1 | 0.79 | 0.84 |
| A2M | 0.79 | 0.81 |
| CCND1 | 0.79 | 0.88 |
| ITIH5 | 0.79 | 0.85 |
| SLCO2B1 | 0.79 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNHG12 | -0.52 | -0.60 |
| COMMD3 | -0.50 | -0.52 |
| EXOSC8 | -0.49 | -0.54 |
| ZNF32 | -0.49 | -0.54 |
| NDUFAF2 | -0.48 | -0.53 |
| RPL27 | -0.48 | -0.60 |
| RPL31 | -0.48 | -0.59 |
| SCNM1 | -0.48 | -0.50 |
| RPS23 | -0.47 | -0.58 |
| RPS27A | -0.47 | -0.61 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF12 | DKFZp686O2372 | KIAA0382 | LARG | PRO2792 | Rho guanine nucleotide exchange factor (GEF) 12 | Biochemical Activity Phenotypic Enhancement | BioGRID | 12515866 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 11071635 |11825908 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | - | HPRD,BioGRID | 12515866 |
| GNA13 | G13 | MGC46138 | guanine nucleotide binding protein (G protein), alpha 13 | Phenotypic Enhancement | BioGRID | 12515866 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9178903 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9473212 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7526158 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | Affinity Capture-Western | BioGRID | 11071635 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Two-hybrid | BioGRID | 9178903 |
| PIK3R3 | DKFZp686P05226 | FLJ41892 | p55 | p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | - | HPRD | 9178903 |
| PLK4 | SAK | STK18 | polo-like kinase 4 (Drosophila) | - | HPRD | 11489907 |
| PRLR | hPRLrI | prolactin receptor | - | HPRD,BioGRID | 11328862 |
| PTPN21 | PTPD1 | PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 | - | HPRD | 11013262 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Phenotypic Enhancement | BioGRID | 11328862 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Reconstituted Complex | BioGRID | 12515866 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 9341160 |
| STAP1 | BRDG1 | STAP-1 | signal transducing adaptor family member 1 | - | HPRD | 10518561 |11716489 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 11328862 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD | 7651724 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD,BioGRID | 8892607 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS STIMULATED DN | 11 | 9 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 6 | 75 | 48 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |