Gene Page: TEP1
Summary ?
| GeneID | 7011 |
| Symbol | TEP1 |
| Synonyms | TLP1|TP1|TROVE1|VAULT2|p240 |
| Description | telomerase associated protein 1 |
| Reference | MIM:601686|HGNC:HGNC:11726|Ensembl:ENSG00000129566|HPRD:03404|Vega:OTTHUMG00000029515 |
| Gene type | protein-coding |
| Map location | 14q11.2 |
| Pascal p-value | 0.605 |
| Sherlock p-value | 0.077 |
| Fetal beta | -0.38 |
| eGene | Myers' cis & trans Meta |
| Support | CompositeSet Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEP1 | chr14 | 20876235 | C | T | NM_007110 | p.122V>M | missense | Schizophrenia | DNM:Xu_2012 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2289256 | chr4 | 187089132 | TEP1 | 7011 | 0.2 | trans | ||
| rs721238 | chr11 | 41679262 | TEP1 | 7011 | 0.16 | trans | ||
| rs9306764 | chrX | 22142353 | TEP1 | 7011 | 0.07 | trans |
Section II. Transcriptome annotation
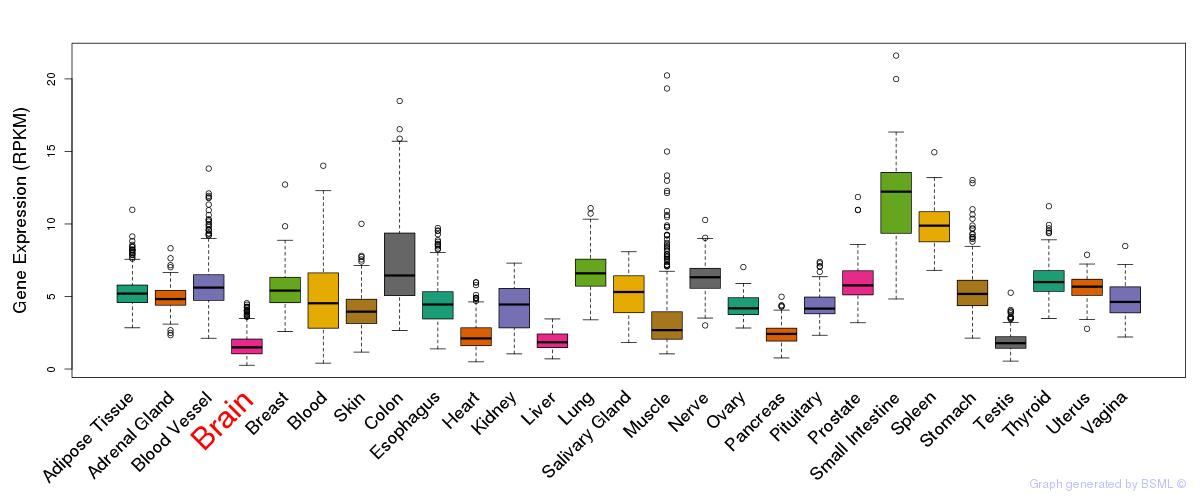
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYTL1 | 0.67 | 0.38 |
| FAM70A | 0.65 | 0.44 |
| LRMP | 0.63 | 0.57 |
| SCN11A | 0.61 | 0.24 |
| KLK7 | 0.59 | 0.61 |
| MATN2 | 0.57 | 0.55 |
| BAIAP3 | 0.57 | 0.68 |
| TJP3 | 0.55 | 0.45 |
| CYP26B1 | 0.55 | 0.47 |
| SLC38A8 | 0.55 | 0.52 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCNI | -0.15 | -0.21 |
| ZNF124 | -0.15 | -0.27 |
| IGFBP2 | -0.15 | -0.35 |
| KIAA1949 | -0.15 | -0.32 |
| BZW2 | -0.15 | -0.23 |
| C5orf13 | -0.14 | -0.22 |
| ISLR2 | -0.14 | -0.20 |
| ZNF695 | -0.14 | -0.26 |
| SH3BP2 | -0.14 | -0.39 |
| TUBB2B | -0.14 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003720 | telomerase activity | IDA | 12135483 | |
| GO:0003723 | RNA binding | ISS | - | |
| GO:0005524 | ATP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000722 | telomere maintenance via recombination | IDA | 10810353 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000781 | chromosome, telomeric region | IEA | - | |
| GO:0005625 | soluble fraction | ISS | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005694 | chromosome | IEA | - | |
| GO:0005697 | telomerase holoenzyme complex | IDA | 12135483 | |
| GO:0005737 | cytoplasm | IDA | 7876352 | |
| GO:0016363 | nuclear matrix | IDA | 7876352 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS DN | 66 | 39 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| CONRAD STEM CELL | 39 | 27 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL SCP2 QTL TRANS | 25 | 13 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |