Gene Page: TGFBR2
Summary ?
| GeneID | 7048 |
| Symbol | TGFBR2 |
| Synonyms | AAT3|FAA3|LDS1B|LDS2|LDS2B|MFS2|RIIC|TAAD2|TGFR-2|TGFbeta-RII |
| Description | transforming growth factor beta receptor II |
| Reference | MIM:190182|HGNC:HGNC:11773|Ensembl:ENSG00000163513|HPRD:01823|Vega:OTTHUMG00000130569 |
| Gene type | protein-coding |
| Map location | 3p22 |
| Pascal p-value | 0.669 |
| Fetal beta | -0.929 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
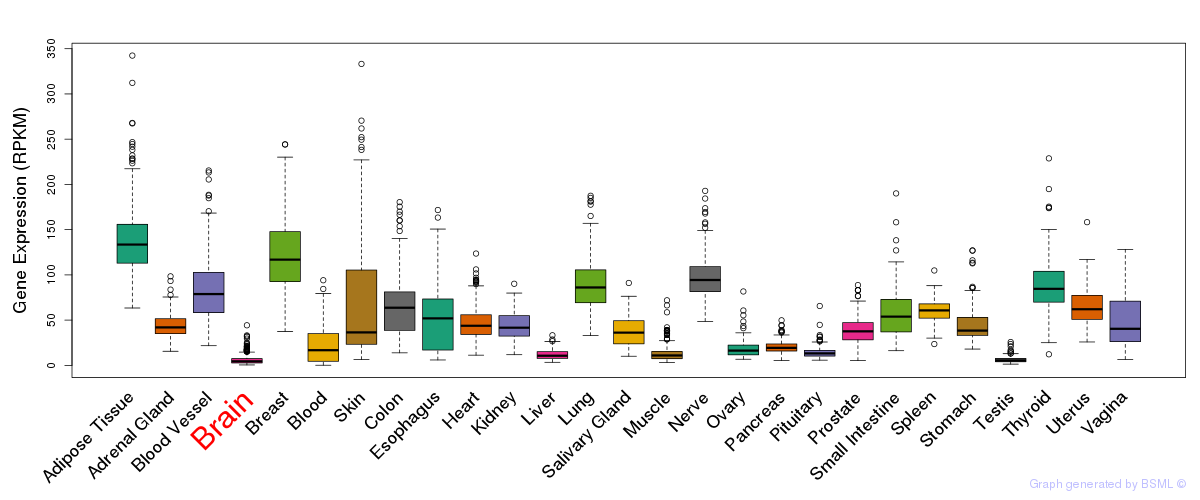
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| INSR | 0.90 | 0.91 |
| BACH2 | 0.89 | 0.88 |
| SYT6 | 0.88 | 0.91 |
| LZTS1 | 0.88 | 0.91 |
| ZNF462 | 0.88 | 0.84 |
| SRGAP1 | 0.87 | 0.80 |
| PLXNA2 | 0.87 | 0.89 |
| RRP1B | 0.87 | 0.90 |
| SRGAP2 | 0.87 | 0.91 |
| SLC22A23 | 0.87 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.61 | -0.82 |
| SERPINB6 | -0.60 | -0.67 |
| HSD17B14 | -0.60 | -0.75 |
| MT-CO2 | -0.60 | -0.82 |
| C5orf53 | -0.59 | -0.69 |
| FXYD1 | -0.59 | -0.81 |
| AIFM3 | -0.59 | -0.71 |
| AF347015.27 | -0.58 | -0.78 |
| S100B | -0.58 | -0.77 |
| AF347015.33 | -0.58 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005024 | transforming growth factor beta receptor activity | IDA | 7852346 |18453574 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004702 | receptor signaling protein serine/threonine kinase activity | IEA | - | |
| GO:0005539 | glycosaminoglycan binding | IDA | 7852346 | |
| GO:0005515 | protein binding | IPI | 8242742 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0050431 | transforming growth factor beta binding | IDA | 7852346 |18243111 |18453574 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | ISS | Brain (GO term level: 7) | - |
| GO:0043011 | myeloid dendritic cell differentiation | ISS | dendrite (GO term level: 10) | - |
| GO:0001570 | vasculogenesis | ISS | - | |
| GO:0001569 | patterning of blood vessels | ISS | - | |
| GO:0001568 | blood vessel development | TAS | 10092230 | |
| GO:0002651 | positive regulation of tolerance induction to self antigen | ISS | - | |
| GO:0002663 | positive regulation of B cell tolerance induction | ISS | - | |
| GO:0002666 | positive regulation of T cell tolerance induction | ISS | - | |
| GO:0002053 | positive regulation of mesenchymal cell proliferation | ISS | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | IDA | 18453574 | |
| GO:0007507 | heart development | ISS | - | |
| GO:0051138 | positive regulation of NK T cell differentiation | ISS | - | |
| GO:0048701 | embryonic cranial skeleton morphogenesis | ISS | - | |
| GO:0032147 | activation of protein kinase activity | ISS | - | |
| GO:0035162 | embryonic hemopoiesis | ISS | - | |
| GO:0060021 | palate development | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043235 | receptor complex | IDA | Neurotransmitter (GO term level: 4) | 8774881 |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009897 | external side of plasma membrane | IDA | 18453574 | |
| GO:0005886 | plasma membrane | EXP | 9865696 |11100470 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVRL1 | ACVRLK1 | ALK-1 | ALK1 | HHT | HHT2 | ORW2 | SKR3 | TSR-I | activin A receptor type II-like 1 | - | HPRD | 10716993 |
| AP2B1 | ADTB2 | AP105B | AP2-BETA | CLAPB1 | DKFZp781K0743 | adaptor-related protein complex 2, beta 1 subunit | Affinity Capture-Western Two-hybrid | BioGRID | 12429842 |
| ARHGEF6 | COOL2 | Cool-2 | KIAA0006 | MRX46 | PIXA | alpha-PIX | alphaPIX | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | T-beta-RII interacts with alpha-PIX. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse alpha-PIX. | BIND | 15761153 |
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | T-beta-RII interacts with Beta-PIX. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse Beta-PIX. | BIND | 15761153 |
| CCNB1 | CCNB | cyclin B1 | - | HPRD,BioGRID | 9926943 |
| CCNB2 | HsT17299 | cyclin B2 | - | HPRD,BioGRID | 9926943 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD | 12145287 |
| CLU | AAG4 | APOJ | CLI | KUB1 | MGC24903 | SGP-2 | SGP2 | SP-40 | TRPM-2 | TRPM2 | clusterin | - | HPRD,BioGRID | 8555189 |
| CLU | AAG4 | APOJ | CLI | KUB1 | MGC24903 | SGP-2 | SGP2 | SP-40 | TRPM-2 | TRPM2 | clusterin | ApoJ interacts with TGFBR2. | BIND | 8555189 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 12000714 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD,BioGRID | 11387212 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD | 11483955 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | - | HPRD | 7566156 |11781097 |
| ENG | CD105 | END | FLJ41744 | HHT1 | ORW | ORW1 | endoglin | - | HPRD,BioGRID | 9872992 |
| FLI1 | EWSR2 | SIC-1 | Friend leukemia virus integration 1 | FLI1 (EWS-FLI1) interacts with the TGFBR2 promoter. | BIND | 15735734 |
| KCNK18 | K2p18.1 | TRESK | TRESK-2 | TRESK2 | TRIK | potassium channel, subfamily K, member 18 | T-beta-RII interacts with TRIK. This interaction was modeled on a demonstrated interaction between human T-beta-RII and TRIK from an unspecified species. | BIND | 15761153 |
| OSR1 | ODD | odd-skipped related 1 (Drosophila) | T-beta-RII interacts with OSR1. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse OSR1. | BIND | 15761153 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | T-beta-RII interacts with PAK1. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse PAK1. | BIND | 15761153 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Affinity Capture-Western | BioGRID | 9435577 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Affinity Capture-Western | BioGRID | 9435577 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | cPML interacts with T-beta-RII. | BIND | 15356634 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 15356634 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | T-beta-RII interacts with an unspecific isoform of PPP1C. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse PPP1C. | BIND | 15761153 |
| SCRIB | CRIB1 | SCRB1 | SCRIB1 | Vartul | scribbled homolog (Drosophila) | T-beta-RII interacts with an unspecified isoform of CRIB. This was modeled interaction on a demonstrated interaction between human T-beta-RII and mouse CRIB. | BIND | 15761153 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | T-beta-RII phosphorylates hMAD-3. This interaction was modeled on a demonstrated interaction between T-beta-RII from an unspecified species and human hMAD-3. | BIND | 8774881 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | T-beta-RII phosphorylates hMAD-4. This interaction was modeled on a demonstrated interaction between T-beta-RII from an unspecified species and human hMAD-4. | BIND | 8774881 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD,BioGRID | 11163210 |
| SMURF2 | DKFZp686F0270 | MGC138150 | SMAD specific E3 ubiquitin protein ligase 2 | Affinity Capture-Western | BioGRID | 11163210 |
| SNX6 | MGC3157 | MSTP010 | TFAF2 | sorting nexin 6 | - | HPRD,BioGRID | 11279102 |
| STRAP | MAWD | PT-WD | UNRIP | serine/threonine kinase receptor associated protein | - | HPRD,BioGRID | 9856985 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | Affinity Capture-Western | BioGRID | 11157754 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD | 7935686 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | TGF-beta-1 interacts with TGF-beta-R-II. This interaction was modeled on a demonstrated interaction between TGF-beta-1 from an unspecified species and human TGF-beta-R-II. | BIND | 7852346 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | - | HPRD | 12421567 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | - | HPRD,BioGRID | 8106553|11157754 |
| TGFB3 | ARVD | FLJ16571 | TGF-beta3 | transforming growth factor, beta 3 | - | HPRD | 8106553 |11382746 |11850637 |12729750 |
| TGFB3 | ARVD | FLJ16571 | TGF-beta3 | transforming growth factor, beta 3 | Affinity Capture-Western Co-crystal Structure Reconstituted Complex | BioGRID | 9872992 |11157754 |11850637 |12729750 |
| TGFB3 | ARVD | FLJ16571 | TGF-beta3 | transforming growth factor, beta 3 | TGF-beta-3 interacts with TGF-beta-R-II. This interaction was modeled on a demonstrated interaction between TGF-beta-3 from an unspecified species and human TGF-beta-R-II. | BIND | 7852346 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD,BioGRID | 7890683 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | T-beta-R1 interacts with T-beta-R2. This interaction was modeled on a demonstrated interaction between mouse T-beta-R1 and mouse T-beta-R2. | BIND | 14612425 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD,BioGRID | 7890683 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | T-beta-RII autophosphorylates. This interaction was modeled on a demonstrated interaction of T-beta-RII from an unspecified species. | BIND | 8774881 |
| TGFBR3 | BGCAN | betaglycan | transforming growth factor, beta receptor III | Affinity Capture-Western | BioGRID | 8106553 |
| TGFBR3 | BGCAN | betaglycan | transforming growth factor, beta receptor III | - | HPRD | 11323414 |
| TGFBRAP1 | TRAP-1 | TRAP1 | transforming growth factor, beta receptor associated protein 1 | - | HPRD | 11278302 |
| TSSK1B | FKSG81 | SPOGA4 | STK22D | TSSK1 | testis-specific serine kinase 1B | T-beta-RII interacts with STK22D. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse STK22D. | BIND | 15761153 |
| ZFYVE9 | MADHIP | NSP | SARA | SMADIP | zinc finger, FYVE domain containing 9 | - | HPRD | 9865696 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOB1 PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| SHI SPARC TARGETS UP | 24 | 17 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM3 | 70 | 37 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN T LYMPHOCYTE DN | 37 | 29 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL CARCINOMA VS ADENOMA DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO RADIATION THERAPY | 32 | 20 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PLURIPOTENT STATE DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND HD MTX DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| LOPEZ TRANSLATION VIA FN1 SIGNALING | 35 | 21 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 266 | 272 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-142-5p | 270 | 276 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-145 | 2194 | 2201 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-153 | 2439 | 2445 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 268 | 275 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-186 | 2338 | 2344 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-19 | 265 | 271 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-204/211 | 2324 | 2331 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-21 | 206 | 212 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-219 | 474 | 481 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-23 | 2383 | 2390 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-320 | 1762 | 1768 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-323 | 2383 | 2389 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-370 | 62 | 69 | 1A,m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-378 | 227 | 233 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-410 | 310 | 316 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-9 | 47 | 54 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.