Gene Page: ACTG1
Summary ?
| GeneID | 71 |
| Symbol | ACTG1 |
| Synonyms | ACT|ACTG|BRWS2|DFNA20|DFNA26|HEL-176 |
| Description | actin gamma 1 |
| Reference | MIM:102560|HGNC:HGNC:144|Ensembl:ENSG00000184009|HPRD:00017|Vega:OTTHUMG00000177955 |
| Gene type | protein-coding |
| Map location | 17q25 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 0.996 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.142:DS1_beta=0.049700:DS2_p=8.57e-01:DS2_beta=0.009:DS2_FDR=9.36e-01 |
| Fetal beta | 1.813 |
| Support | STRUCTURAL PLASTICITY |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.083 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
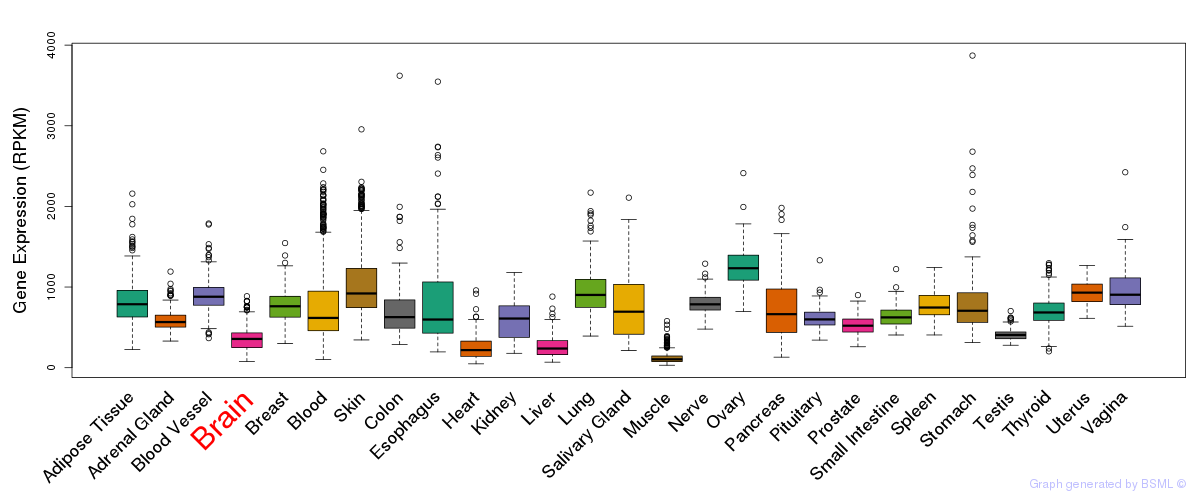
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | IC | 16130169 | |
| GO:0042802 | identical protein binding | IPI | 16189514 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0006928 | cell motion | TAS | 16130169 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | TAS | 16130169 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | Two-hybrid | BioGRID | 16189514 |
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Two-hybrid | BioGRID | 16189514 |
| ANXA5 | ANX5 | ENX2 | PP4 | annexin A5 | - | HPRD | 10903505 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD,BioGRID | 10958671 |
| CAP1 | CAP | CAP1-PEN | CAP, adenylate cyclase-associated protein 1 (yeast) | Affinity Capture-Western Two-hybrid | BioGRID | 8761950 |
| CAP2 | - | CAP, adenylate cyclase-associated protein, 2 (yeast) | - | HPRD,BioGRID | 8761950 |
| CAPZA3 | CAPPA3 | Gsg3 | capping protein (actin filament) muscle Z-line, alpha 3 | - | HPRD | 12029070 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western | BioGRID | 9535896 |
| CFL1 | CFL | cofilin 1 (non-muscle) | Two-hybrid | BioGRID | 16189514 |
| CFL2 | NEM7 | cofilin 2 (muscle) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | Affinity Capture-Western | BioGRID | 12835311 |
| DNASE1 | DKFZp686H0155 | DNL1 | DRNI | FLJ38093 | FLJ44902 | deoxyribonuclease I | - | HPRD | 11474115 |
| DSTN | ACTDP | ADF | bA462D18.2 | destrin (actin depolymerizing factor) | - | HPRD | 8399167 |
| DSTN | ACTDP | ADF | bA462D18.2 | destrin (actin depolymerizing factor) | Two-hybrid | BioGRID | 16189514 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Two-hybrid | BioGRID | 16169070 |
| EPS8L1 | DRC3 | EPS8R1 | FLJ20258 | MGC23164 | MGC4642 | PP10566 | EPS8-like 1 | Affinity Capture-Western | BioGRID | 14565974 |
| EPS8L2 | EPS8R2 | FLJ16738 | FLJ21935 | FLJ22171 | MGC126530 | MGC3088 | EPS8-like 2 | Affinity Capture-Western | BioGRID | 14565974 |
| EPS8L3 | EPS8R3 | FLJ21522 | MGC16817 | EPS8-like 3 | Affinity Capture-Western | BioGRID | 14565974 |
| FEZ1 | - | fasciculation and elongation protein zeta 1 (zygin I) | Affinity Capture-Western | BioGRID | 12874605 |
| FHOD1 | FHOS | formin homology 2 domain containing 1 | Affinity Capture-Western | BioGRID | 14576350 |
| LGALS13 | GAL13 | PLAC8 | PP13 | lectin, galactoside-binding, soluble, 13 | - | HPRD | 15009185 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | Affinity Capture-MS | BioGRID | 10772929 |
| MYO1A | BBMI | DFNA48 | MIHC | MYHL | myosin IA | - | HPRD | 11474115 |
| PFN2 | D3S1319E | PFL | profilin 2 | Two-hybrid | BioGRID | 16169070 |
| PLG | DKFZp779M0222 | plasminogen | PLG interacts with ACTG1 (G-actin). This interaction was modelled on a demonstrated interaction between human PLG and bovine ACTG1. | BIND | 15746964 |
| PPP1R9A | FLJ20068 | KIAA1222 | NRB1 | NRBI | Neurabin-I | protein phosphatase 1, regulatory (inhibitor) subunit 9A | - | HPRD | 9362513 |12052877 |
| PRSS23 | MGC5107 | SIG13 | SPUVE | ZSIG13 | protease, serine, 23 | Two-hybrid | BioGRID | 16169070 |
| RBX1 | BA554C12.1 | MGC13357 | MGC1481 | RNF75 | ROC1 | ring-box 1 | Affinity Capture-MS | BioGRID | 12481031 |
| SCIN | KIAA1905 | scinderin | - | HPRD | 1331119 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | Two-hybrid | BioGRID | 16169070 |
| ST3GAL3 | SIAT6 | ST3GALII | ST3GalIII | ST3N | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 | Two-hybrid | BioGRID | 16169070 |
| TMSB4X | FX | PTMB4 | TB4X | TMSB4 | thymosin beta 4, X-linked | - | HPRD | 8617195 |8630056 |9153421 |10848969 |15163409 |
| TMSB4X | FX | PTMB4 | TB4X | TMSB4 | thymosin beta 4, X-linked | Co-crystal Structure Reconstituted Complex | BioGRID | 8617195 |15163409 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | WAVE-1 interacts with G-actin. This interaction was modeled on a demonstrated interaction between human WAVE-1 and rat G-actin. | BIND | 10970852 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG VIBRIO CHOLERAE INFECTION | 56 | 32 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | 35 | 25 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC UP | 54 | 30 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS NEONATAL | 35 | 25 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| MMS MOUSE LYMPH HIGH 4HRS UP | 36 | 20 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WENG POR DOSAGE | 21 | 9 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 358 | 364 | 1A | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 26 | 32 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-145 | 96 | 103 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-186 | 610 | 616 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-199 | 151 | 157 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-361 | 140 | 146 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-381 | 184 | 190 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-485-5p | 86 | 92 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.