Gene Page: C1QA
Summary ?
| GeneID | 712 |
| Symbol | C1QA |
| Synonyms | - |
| Description | complement component 1, q subcomponent, A chain |
| Reference | MIM:120550|HGNC:HGNC:1241|Ensembl:ENSG00000173372|HPRD:00393|Vega:OTTHUMG00000002893 |
| Gene type | protein-coding |
| Map location | 1p36.12 |
| Pascal p-value | 0.696 |
| Sherlock p-value | 0.258 |
| Fetal beta | -1.492 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7060495 | chrX | 36100343 | C1QA | 712 | 0.12 | trans |
Section II. Transcriptome annotation
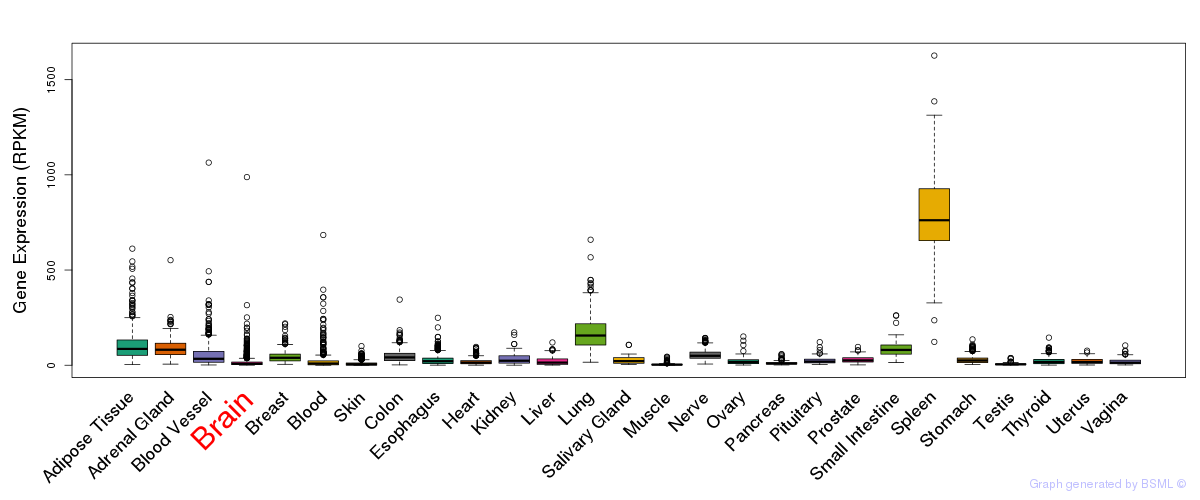
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1R | 0.78 | 0.81 |
| MR1 | 0.70 | 0.73 |
| HIBADH | 0.70 | 0.72 |
| CTSL1 | 0.70 | 0.77 |
| DTNA | 0.70 | 0.77 |
| ITM2B | 0.69 | 0.72 |
| IL1R1 | 0.69 | 0.74 |
| ANXA7 | 0.68 | 0.73 |
| ME1 | 0.68 | 0.74 |
| PGM1 | 0.68 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZBTB12 | -0.53 | -0.46 |
| SH2B2 | -0.53 | -0.56 |
| H2AFY2 | -0.53 | -0.44 |
| C9orf7 | -0.53 | -0.46 |
| TUBB2B | -0.52 | -0.46 |
| MARCKSL1 | -0.52 | -0.47 |
| FAM110A | -0.52 | -0.44 |
| KIAA1949 | -0.52 | -0.44 |
| GPC2 | -0.52 | -0.48 |
| SOX11 | -0.52 | -0.46 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APCS | MGC88159 | PTX2 | SAP | amyloid P component, serum | - | HPRD | 8417122 |
| C1QB | - | complement component 1, q subcomponent, B chain | - | HPRD | 12960167 |
| C1QC | C1Q-C | C1QG | FLJ27103 | complement component 1, q subcomponent, C chain | - | HPRD | 12960167 |
| C1R | - | complement component 1, r subcomponent | - | HPRD,BioGRID | 10878362 |
| C1S | FLJ44757 | complement component 1, s subcomponent | - | HPRD,BioGRID | 10878362 |
| CD93 | C1QR1 | C1qR(P) | C1qRP | CDw93 | MXRA4 | dJ737E23.1 | CD93 molecule | - | HPRD,BioGRID | 9476117 |
| COL2A1 | ANFH | AOM | COL11A3 | MGC131516 | SEDC | collagen, type II, alpha 1 | - | HPRD,BioGRID | 8778019 |
| CR1 | C3BR | CD35 | KN | complement component (3b/4b) receptor 1 (Knops blood group) | - | HPRD,BioGRID | 10528211 |
| CRP | MGC149895 | MGC88244 | PTX1 | C-reactive protein, pentraxin-related | Reconstituted Complex | BioGRID | 10490997 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD,BioGRID | 1431141 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD,BioGRID | 6981115 |
| HRG | DKFZp779H1622 | HPRG | HRGP | histidine-rich glycoprotein | - | HPRD,BioGRID | 9184145 |
| PTX3 | TNFAIP5 | TSG-14 | pentraxin-related gene, rapidly induced by IL-1 beta | - | HPRD,BioGRID | 12645945 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CLASSIC PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA COMP PATHWAY | 19 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME COMPLEMENT CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CREATION OF C4 AND C2 ACTIVATORS | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME INITIAL TRIGGERING OF COMPLEMENT | 16 | 10 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU UP | 53 | 37 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M | 59 | 36 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP | 76 | 46 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING UP | 55 | 39 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| HECKER IFNB1 TARGETS | 95 | 54 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |