Gene Page: TNNT1
Summary ?
| GeneID | 7138 |
| Symbol | TNNT1 |
| Synonyms | ANM|NEM5|STNT|TNT|TNTS |
| Description | troponin T1, slow skeletal type |
| Reference | MIM:191041|HGNC:HGNC:11948|Ensembl:ENSG00000105048|HPRD:01841|Vega:OTTHUMG00000180542 |
| Gene type | protein-coding |
| Map location | 19q13.4 |
| Pascal p-value | 0.009 |
| Sherlock p-value | 0.893 |
| Fetal beta | -2.257 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19393762 | 19 | 55661872 | TNNT1 | 3.61E-6 | -0.446 | 0.009 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7258577 | chr19 | 55186254 | TNNT1 | 7138 | 0.16 | cis | ||
| rs17714747 | chr2 | 205010190 | TNNT1 | 7138 | 0.15 | trans | ||
| rs9313483 | chr5 | 169448229 | TNNT1 | 7138 | 0.05 | trans |
Section II. Transcriptome annotation
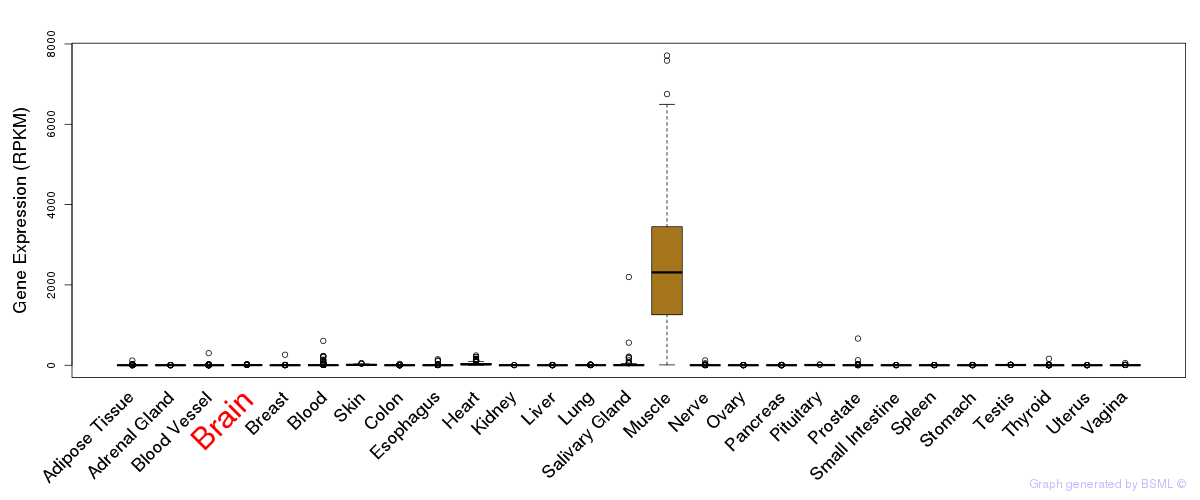
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRPF39 | 0.87 | 0.89 |
| PEX1 | 0.87 | 0.88 |
| CCDC82 | 0.86 | 0.84 |
| WDR75 | 0.86 | 0.86 |
| SLC35A1 | 0.85 | 0.84 |
| TUBE1 | 0.85 | 0.82 |
| SMC6 | 0.85 | 0.85 |
| MFN1 | 0.84 | 0.84 |
| ZNF187 | 0.84 | 0.86 |
| UTP6 | 0.84 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.65 | -0.74 |
| AF347015.31 | -0.65 | -0.75 |
| AF347015.27 | -0.64 | -0.72 |
| MT-CYB | -0.63 | -0.73 |
| AF347015.8 | -0.63 | -0.74 |
| HLA-F | -0.62 | -0.66 |
| AF347015.33 | -0.62 | -0.71 |
| AF347015.15 | -0.61 | -0.73 |
| IFI27 | -0.61 | -0.73 |
| AF347015.2 | -0.60 | -0.74 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| PLEKHF1 | APPD | MGC4090 | PHAFIN1 | ZFYVE15 | pleckstrin homology domain containing, family F (with FYVE domain) member 1 | Two-hybrid | BioGRID | 16189514 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | Two-hybrid | BioGRID | 16189514 |
| PRKG1 | CGKI | DKFZp686K042 | FLJ36117 | MGC71944 | PGK | PKG | PRKG1B | PRKGR1B | cGKI-BETA | cGKI-alpha | protein kinase, cGMP-dependent, type I | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10601315 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Two-hybrid | BioGRID | 16189514 |
| RP4-691N24.1 | FLJ11792 | KIAA0980 | NLP | dJ691N24.1 | ninein-like | Two-hybrid | BioGRID | 16189514 |
| TNNC1 | CMD1Z | TNC | TNNC | troponin C type 1 (slow) | Two-hybrid | BioGRID | 9448267 |
| TNNI3 | CMD2A | CMH7 | MGC116817 | TNNC1 | cTnI | troponin I type 3 (cardiac) | - | HPRD | 11904166 |
| TNNT1 | ANM | FLJ98147 | MGC104241 | STNT | TNT | TNTS | troponin T type 1 (skeletal, slow) | Two-hybrid | BioGRID | 16189514 |
| TPM1 | C15orf13 | CMD1Y | HTM-alpha | TMSA | tropomyosin 1 (alpha) | Reconstituted Complex | BioGRID | 6822572 |
| ZMYND19 | MIZIP | RP11-48C7.4 | zinc finger, MYND-type containing 19 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME STRIATED MUSCLE CONTRACTION | 27 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MUSCLE CONTRACTION | 48 | 24 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| BHATTACHARYA EMBRYONIC STEM CELL | 89 | 60 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM UP | 38 | 30 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| WANG NFKB TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| CHEMELLO SOLEUS VS EDL MYOFIBERS UP | 35 | 16 | All SZGR 2.0 genes in this pathway |