Gene Page: TP73
Summary ?
| GeneID | 7161 |
| Symbol | TP73 |
| Synonyms | P73 |
| Description | tumor protein p73 |
| Reference | MIM:601990|HGNC:HGNC:12003|Ensembl:ENSG00000078900|HPRD:03587|Vega:OTTHUMG00000000610 |
| Gene type | protein-coding |
| Map location | 1p36.3 |
| Pascal p-value | 0.447 |
| Fetal beta | 0.223 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04137576 | 1 | 3575674 | TP73 | 1.767E-4 | -0.381 | 0.034 | DMG:Wockner_2014 |
| cg24845675 | 1 | 3649971 | TP73 | 2.317E-4 | 0.466 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7370564 | 0 | TP73 | 7161 | 0.18 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
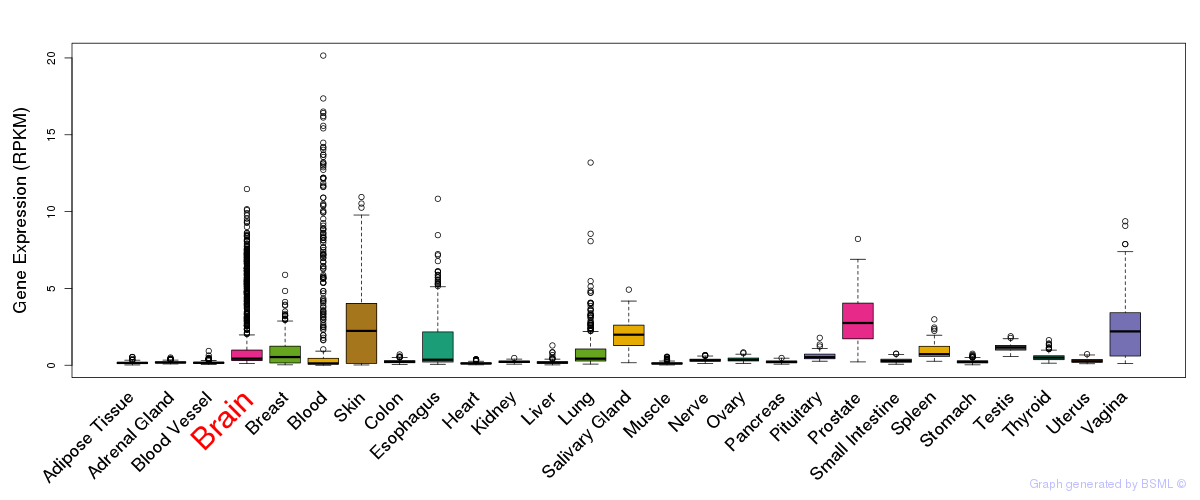
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZCCHC14 | 0.96 | 0.97 |
| CAMSAP1 | 0.96 | 0.98 |
| MAPKBP1 | 0.95 | 0.96 |
| ADD2 | 0.95 | 0.97 |
| XXbac-B461K10.1 | 0.95 | 0.94 |
| SRGAP2 | 0.94 | 0.97 |
| PPP1R10 | 0.94 | 0.95 |
| DIP2C | 0.94 | 0.94 |
| USP42 | 0.94 | 0.96 |
| PIK3C2B | 0.94 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.74 | -0.89 |
| S100B | -0.74 | -0.86 |
| TSC22D4 | -0.74 | -0.80 |
| FXYD1 | -0.73 | -0.86 |
| AF347015.27 | -0.73 | -0.86 |
| MT-CO2 | -0.73 | -0.88 |
| AIFM3 | -0.73 | -0.76 |
| SERPINB6 | -0.72 | -0.78 |
| HEPN1 | -0.72 | -0.75 |
| AF347015.33 | -0.72 | -0.84 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 10391250 |10391251 |
| BAX | BCL2L4 | BCL2-associated X protein | p73-alpha interacts with BAX promoter. | BIND | 15893728 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | p73-alpha binds p53 AIP1 promoter. | BIND | 15175157 |
| CABLES1 | CABLES | FLJ35924 | HsT2563 | IK3-1 | Cdk5 and Abl enzyme substrate 1 | - | HPRD | 11706030 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p73-beta interacts with p21 promoter. This interaction was modeled on a demonstrated interaction between p73-beta from an unspecified species and human p21 promoter. | BIND | 15893728 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p73-alpha interacts with p21 promoter. | BIND | 15893728 |
| CEBPZ | CBF2 | HSP-CBF | NOC1 | CCAAT/enhancer binding protein (C/EBP), zeta | - | HPRD,BioGRID | 12534345 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | - | HPRD,BioGRID | 10961991 |
| DENND4A | FLJ33949 | IRLB | KIAA0476 | MYCPBP | DENN/MADD domain containing 4A | p73 alpha interacts with c-Myc. | BIND | 12080043 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p73-alpha interacts with p300.This interaction was modeled on a demonstrated interaction between human p73-alpha and p300 from an unspecified species. | BIND | 15175157 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | RACK1 interacts with p73. | BIND | 12761493 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 12761493 |
| HECW2 | DKFZp686M17164 | NEDL2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | - | HPRD | 12890487 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD,BioGRID | 11925430 |
| HIPK3 | DYRK6 | FIST3 | PKY | YAK1 | homeodomain interacting protein kinase 3 | Two-hybrid | BioGRID | 10961991 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | p73alpha interacts with HMGB1. | BIND | 11748232 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | p73-beta interacts with HMGB1. | BIND | 11748232 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 11748232 |
| ITCH | AIF4 | AIP4 | NAPP1 | dJ468O1.1 | itchy E3 ubiquitin protein ligase homolog (mouse) | p73-alpha interacts with Itch. | BIND | 15678106 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 10207051 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 12080043 |
| NEDD4 | KIAA0093 | MGC176705 | NEDD4-1 | RPF1 | neural precursor cell expressed, developmentally down-regulated 4 | p73-alpha interacts with an unspecified isoform of Nedd4. | BIND | 15678106 |
| NFYB | CBF-A | CBF-B | HAP3 | NF-YB | nuclear transcription factor Y, beta | - | HPRD,BioGRID | 12167641 |
| NFYC | CBF-C | CBFC | DKFZp667G242 | FLJ45775 | H1TF2A | HAP5 | HSM | NF-YC | nuclear transcription factor Y, gamma | Affinity Capture-Western | BioGRID | 12167641 |
| P53AIP1 | - | p53-regulated apoptosis-inducing protein 1 | p73-alpha interacts with p53AIP1 promoter. | BIND | 15893728 |
| PFDN5 | MGC5329 | MGC71907 | MM-1 | MM1 | PFD5 | prefoldin subunit 5 | MM1 interacts with p73-alpha. | BIND | 11844794 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Pin1 interacts with phosphorylated p73-alpha | BIND | 15175157 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Pin1 interacts with p73-beta. | BIND | 15175157 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 11832207 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western | BioGRID | 12761493 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Two-hybrid | BioGRID | 10961991 |
| TDG | - | thymine-DNA glycosylase | Two-hybrid | BioGRID | 10961991 |
| TP73 | P73 | tumor protein p73 | Two-hybrid | BioGRID | 10961991 |
| TP73 | P73 | tumor protein p73 | p73 alpha interacts with itself. | BIND | 11389695 |
| TP73 | P73 | tumor protein p73 | p73 splicing variants interact with each other. | BIND | 11389695 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 10961991 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 11278685 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | p73-alpha interacts with YAP. | BIND | 15893728 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK14 TARGETS | 10 | 10 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| OHM EMBRYONIC CARCINOMA DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| LIU TOPBP1 TARGETS | 16 | 8 | All SZGR 2.0 genes in this pathway |