Gene Page: TUBG1
Summary ?
| GeneID | 7283 |
| Symbol | TUBG1 |
| Synonyms | CDCBM4|GCP-1|TUBG|TUBGCP1 |
| Description | tubulin gamma 1 |
| Reference | MIM:191135|HGNC:HGNC:12417|Ensembl:ENSG00000131462|HPRD:01853|Vega:OTTHUMG00000180277 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.062 |
| Sherlock p-value | 0.029 |
| Fetal beta | -0.664 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0833 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4854166 | chr2 | 3297005 | TUBG1 | 7283 | 0.13 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
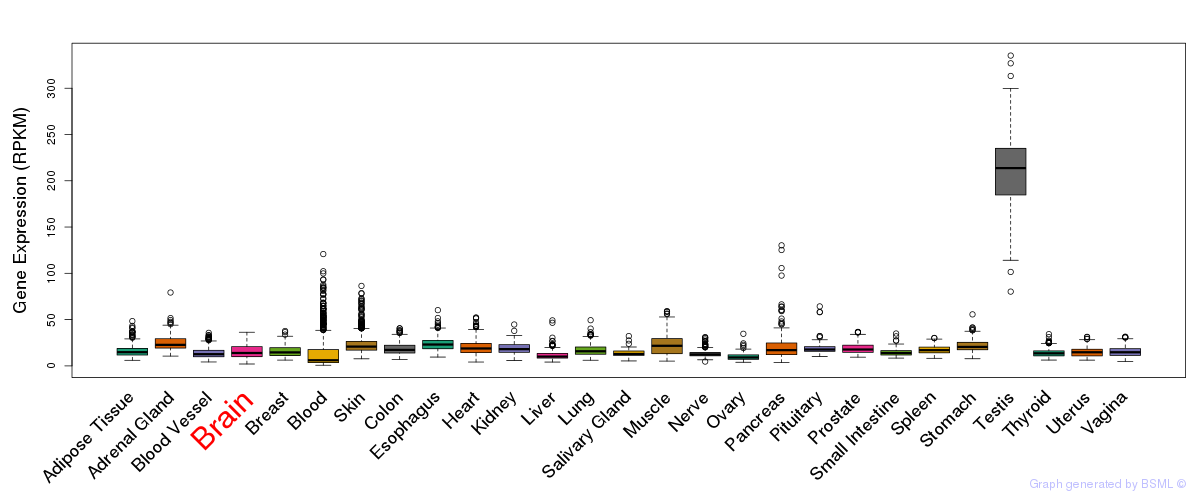
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPS27 | 0.91 | 0.90 |
| KPNA4 | 0.90 | 0.91 |
| NPEPPS | 0.90 | 0.89 |
| GOLGA5 | 0.89 | 0.89 |
| ATF2 | 0.89 | 0.90 |
| CUL1 | 0.89 | 0.89 |
| SPOP | 0.89 | 0.89 |
| VPS37A | 0.89 | 0.89 |
| ARMC1 | 0.89 | 0.90 |
| PSMD11 | 0.89 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.81 | -0.82 |
| AF347015.31 | -0.80 | -0.81 |
| AF347015.8 | -0.79 | -0.81 |
| MT-CYB | -0.78 | -0.80 |
| AF347015.21 | -0.77 | -0.79 |
| AF347015.2 | -0.77 | -0.81 |
| AF347015.33 | -0.77 | -0.78 |
| AF347015.27 | -0.76 | -0.78 |
| AF347015.15 | -0.75 | -0.79 |
| AF347015.26 | -0.75 | -0.78 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 9566969 |17286961 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 1904010 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000212 | meiotic spindle organization | ISS | - | |
| GO:0000226 | microtubule cytoskeleton organization | TAS | 1904010 | |
| GO:0051258 | protein polymerization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000242 | pericentriolar material | IEA | - | |
| GO:0000794 | condensed nuclear chromosome | ISS | - | |
| GO:0000930 | gamma-tubulin complex | TAS | 9566969 | |
| GO:0005816 | spindle pole body | TAS | 1904010 | |
| GO:0005827 | polar microtubule | IDA | 9566969 | |
| GO:0005881 | cytoplasmic microtubule | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 9566969 | |
| GO:0005737 | cytoplasm | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9789027 |11691781 |
| CENPJ | BM032 | CPAP | LAP | LIP1 | MCPH6 | MGC131581 | MGC131582 | MGC142222 | MGC142224 | centromere protein J | Affinity Capture-Western | BioGRID | 11003675 |
| MARK4 | FLJ90097 | KIAA1860 | MARKL1 | Nbla00650 | MAP/microtubule affinity-regulating kinase 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14594945 |
| NDE1 | FLJ20101 | HOM-TES-87 | NUDE | NUDE1 | nudE nuclear distribution gene E homolog 1 (A. nidulans) | Affinity Capture-Western | BioGRID | 11163258 |
| NUP85 | FLJ12549 | Nup75 | nucleoporin 85kDa | - | HPRD | 7592789 |
| PCNT | KEN | MOPD2 | PCN | PCNT2 | PCNTB | PCTN2 | SCKL4 | pericentrin | - | HPRD | 12221128 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 7592789 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 10840040 |11950600 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12639940 |
| RACGAP1 | HsCYK-4 | ID-GAP | MgcRacGAP | Rac GTPase activating protein 1 | - | HPRD | 11942621 |
| RASSF1 | 123F2 | NORE2A | RASSF1A | RDA32 | REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | - | HPRD,BioGRID | 14603253 |
| RNF19A | DKFZp566B1346 | DORFIN | RNF19 | ring finger protein 19A | - | HPRD | 11237715 |
| SPATC1 | MGC61633 | SPATA15 | SPERIOLIN | spermatogenesis and centriole associated 1 | Affinity Capture-Western | BioGRID | 15280373 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | - | HPRD | 11082048 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 11082048 |
| TUBGCP3 | GCP3 | SPBC98 | Spc98p | tubulin, gamma complex associated protein 3 | - | HPRD | 9566969 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 26 | 32 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 26 | 32 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 81 | 87 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-199 | 181 | 187 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.