Gene Page: VDR
Summary ?
| GeneID | 7421 |
| Symbol | VDR |
| Synonyms | NR1I1|PPP1R163 |
| Description | vitamin D (1,25- dihydroxyvitamin D3) receptor |
| Reference | MIM:601769|HGNC:HGNC:12679|Ensembl:ENSG00000111424|HPRD:03463|Vega:OTTHUMG00000169890 |
| Gene type | protein-coding |
| Map location | 12q13.11 |
| Pascal p-value | 0.039 |
| Fetal beta | -0.335 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0172 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
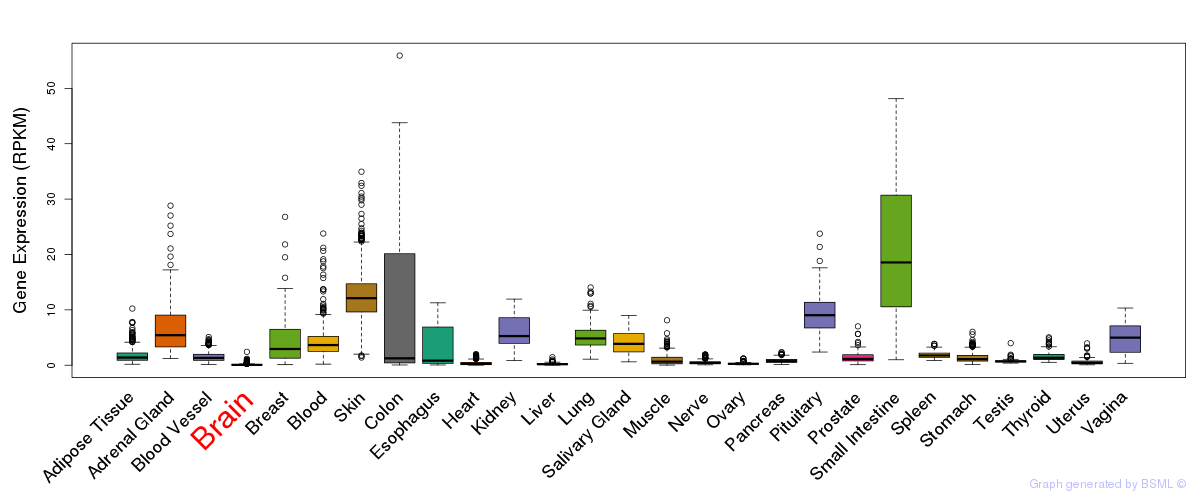
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTMR4 | 0.86 | 0.91 |
| NMNAT2 | 0.85 | 0.90 |
| KIAA0427 | 0.84 | 0.88 |
| KIAA0317 | 0.84 | 0.91 |
| L1CAM | 0.84 | 0.90 |
| KIF3C | 0.83 | 0.90 |
| CNNM4 | 0.83 | 0.89 |
| RAPGEF1 | 0.83 | 0.87 |
| KIAA1409 | 0.83 | 0.88 |
| DIP2C | 0.83 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.68 | -0.81 |
| MT-CO2 | -0.65 | -0.79 |
| AF347015.27 | -0.65 | -0.78 |
| FXYD1 | -0.65 | -0.76 |
| AF347015.8 | -0.65 | -0.79 |
| MT-CYB | -0.64 | -0.77 |
| AF347015.33 | -0.64 | -0.75 |
| HIGD1B | -0.64 | -0.78 |
| CLDN10 | -0.63 | -0.76 |
| COPZ2 | -0.63 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 10866662 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008434 | vitamin D3 receptor activity | TAS | 9070272 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007165 | signal transduction | TAS | 2849209 | |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0016481 | negative regulation of transcription | IDA | 11891224 | |
| GO:0050892 | intestinal absorption | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAG1 | RAP46 | BCL2-associated athanogene | - | HPRD,BioGRID | 10967105 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD,BioGRID | 12837248 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | VDR interacts with hMo15. | BIND | 15249124 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | Reconstituted Complex | BioGRID | 10330159 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | VDR interacts with hTFIIB. | BIND | 15249124 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | - | HPRD,BioGRID | 11358960 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | Rbp2 interacts with VDR. | BIND | 11358960 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 10330159 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10198638 |12837248 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 9653119 |
| MED12 | CAGH45 | HOPA | KIAA0192 | OPA1 | TNRC11 | TRAP230 | mediator complex subunit 12 | Affinity Capture-MS | BioGRID | 10198638 |12837248 |
| MED13 | ARC250 | DRIP250 | HSPC221 | KIAA0593 | THRAP1 | TRAP240 | mediator complex subunit 13 | Affinity Capture-MS | BioGRID | 10198638 |
| MED14 | CRSP150 | CRSP2 | CSRP | CXorf4 | DRIP150 | EXLM1 | MGC104513 | RGR1 | TRAP170 | mediator complex subunit 14 | Affinity Capture-MS | BioGRID | 10198638 |
| MED16 | DRIP92 | THRAP5 | TRAP95 | mediator complex subunit 16 | Affinity Capture-MS | BioGRID | 10198638 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-MS | BioGRID | 10198638 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | Affinity Capture-MS | BioGRID | 10198638 |12837248 |
| MED6 | NY-REN-28 | mediator complex subunit 6 | Affinity Capture-MS | BioGRID | 10198638 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9878542 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | VDR interacts with SRC-1. | BIND | 15249124 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10748178 |12612084 |12837248 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | GRIP1 interacts with VDR. This interaction was modeled on a demonstrated interaction between mouse GRIP1 and human VDR. | BIND | 9920895 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | ACTR interacts with VDR. | BIND | 9267036 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Reconstituted Complex | BioGRID | 12460926 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Reconstituted Complex | BioGRID | 12460926 |
| POU1F1 | GHF-1 | PIT1 | Pit-1 | POU class 1 homeobox 1 | Reconstituted Complex | BioGRID | 11891224 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Reconstituted Complex | BioGRID | 10383413 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Reconstituted Complex | BioGRID | 11358960 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12460926 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12460926 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex Two-hybrid | BioGRID | 9632709 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | VDR interacts with RXRA (RXRalpha) to form a heterodimer. | BIND | 15829977 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | VDR interacts with Smad3. This interaction was modeled on a demonstrated interaction between VDR and Smad3 from unspecified sources. | BIND | 11278756 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-Western | BioGRID | 12837248 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | - | HPRD,BioGRID | 9632709 |12529369 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 11909970 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 12874288 |
| THRAP3 | FLJ22082 | MGC133082 | MGC133083 | TRAP150 | thyroid hormone receptor associated protein 3 | Affinity Capture-MS | BioGRID | 10198638 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD | 9115274 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | Affinity Capture-MS | BioGRID | 12837248 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | - | HPRD,BioGRID | 11980721 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 11719366 |12460926 |14521715 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| LANDIS BREAST CANCER PROGRESSION UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SAENZ DETOX PATHWAY AND CARCINOGENESIS DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 6HR | 40 | 23 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| KOMMAGANI TP63 GAMMA TARGETS | 9 | 7 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR DN | 47 | 31 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2 | 72 | 52 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 8HR | 39 | 31 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MATZUK STEROIDOGENESIS | 7 | 6 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC ENDOCRINE PROGENITOR | 14 | 11 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 5 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| KANG AR TARGETS UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WANG THOC1 TARGETS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| ROLEF GLIS3 TARGETS | 37 | 29 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 362 | 369 | 1A,m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.