Gene Page: VIM
Summary ?
| GeneID | 7431 |
| Symbol | VIM |
| Synonyms | CTRCT30|HEL113 |
| Description | vimentin |
| Reference | MIM:193060|HGNC:HGNC:12692|Ensembl:ENSG00000026025|HPRD:01899|Vega:OTTHUMG00000017744 |
| Gene type | protein-coding |
| Map location | 10p13 |
| Pascal p-value | 0.213 |
| Sherlock p-value | 0.264 |
| Fetal beta | 2.193 |
| eGene | Anterior cingulate cortex BA24 Cortex Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9451867 | chr6 | 92659609 | VIM | 7431 | 0 | trans | ||
| rs2199402 | chr8 | 9201002 | VIM | 7431 | 0.05 | trans | ||
| rs7841407 | chr8 | 9243427 | VIM | 7431 | 0.01 | trans | ||
| rs7007515 | chr8 | 117444193 | VIM | 7431 | 0.15 | trans | ||
| snp_a-2151892 | 0 | VIM | 7431 | 0.15 | trans | |||
| rs17073665 | chr13 | 81541961 | VIM | 7431 | 0.04 | trans | ||
| rs11873703 | chr18 | 61448702 | VIM | 7431 | 0.11 | trans | ||
| rs7084057 | 10 | 17048022 | VIM | ENSG00000026025.9 | 2.49916E-6 | 0.05 | -222236 | gtex_brain_ba24 |
Section II. Transcriptome annotation
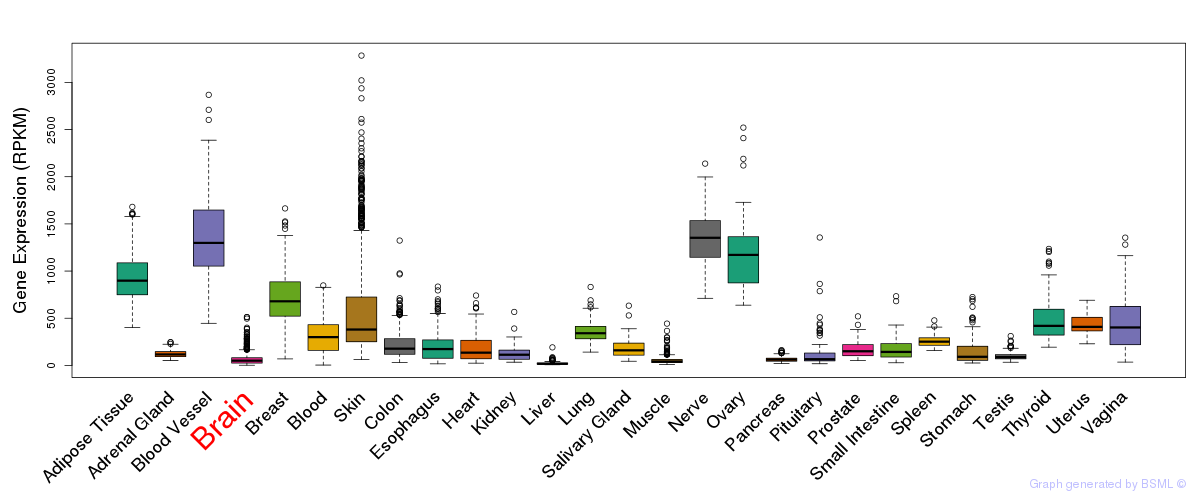
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FGFR1 | 0.68 | 0.58 |
| MTTP | 0.65 | 0.55 |
| VANGL1 | 0.64 | 0.59 |
| TMEM47 | 0.63 | 0.67 |
| PLEKHG1 | 0.62 | 0.54 |
| TNFRSF19 | 0.62 | 0.54 |
| CTNNB1 | 0.61 | 0.66 |
| LATS2 | 0.60 | 0.49 |
| GNS | 0.60 | 0.75 |
| MDFIC | 0.60 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.32 | -0.41 |
| MT-CO2 | -0.32 | -0.41 |
| AF347015.21 | -0.31 | -0.42 |
| HIGD1B | -0.30 | -0.40 |
| AC098691.2 | -0.30 | -0.48 |
| FXYD1 | -0.30 | -0.34 |
| CXCL14 | -0.30 | -0.34 |
| AF347015.33 | -0.29 | -0.34 |
| AF347015.2 | -0.29 | -0.35 |
| S100A13 | -0.29 | -0.36 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IDA | 11889032 | |
| GO:0005515 | protein binding | IPI | 11827972 | |
| GO:0005200 | structural constituent of cytoskeleton | IDA | 11889032 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006928 | cell motion | TAS | 16130169 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0045103 | intermediate filament-based process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | TAS | 16130169 | |
| GO:0005882 | intermediate filament | IDA | 11889032 | |
| GO:0005737 | cytoplasm | IDA | 9150946 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABLIM1 | ABLIM | DKFZp781D0148 | FLJ14564 | KIAA0059 | LIMAB1 | LIMATIN | MGC1224 | actin binding LIM protein 1 | Two-hybrid | BioGRID | 16189514 |
| ALS2CR11 | FLJ25351 | FLJ40332 | amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 | Two-hybrid | BioGRID | 16189514 |
| APIP | APIP2 | CGI-29 | MMRP19 | dJ179L10.2 | APAF1 interacting protein | Two-hybrid | BioGRID | 16189514 |
| ARS2 | ASR2 | MGC126427 | arsenate resistance protein 2 | Two-hybrid | BioGRID | 16169070 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Two-hybrid | BioGRID | 16169070 |
| BFSP1 | CP115 | CP94 | FILENSIN | LIFL-H | beaded filament structural protein 1, filensin | - | HPRD,BioGRID | 1478967 |1918147 |
| BHLHB2 | DEC1 | SHARP-2 | STRA13 | Stra14 | bHLHe40 | basic helix-loop-helix domain containing, class B, 2 | Two-hybrid | BioGRID | 16169070 |
| BRD1 | BRL | BRPF1 | BRPF2 | DKFZp686F0325 | bromodomain containing 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| C7orf36 | GK003 | chromosome 7 open reading frame 36 | Two-hybrid | BioGRID | 16169070 |
| CAPN1 | CANP | CANP1 | CANPL1 | muCANP | muCL | calpain 1, (mu/I) large subunit | CAPN1 interacts with VIM. | BIND | 12358155 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Two-hybrid | BioGRID | 16169070 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | - | HPRD,BioGRID | 12003790 |
| CRCT1 | C1orf42 | NICE-1 | cysteine-rich C-terminal 1 | Two-hybrid | BioGRID | 16169070 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD,BioGRID | 14600259 |
| DIS3L2 | FAM6A | FLJ36974 | MGC42174 | DIS3 mitotic control homolog (S. cerevisiae)-like 2 | Two-hybrid | BioGRID | 16169070 |
| DPPA4 | 2410091M23Rik | FLJ10713 | developmental pluripotency associated 4 | Two-hybrid | BioGRID | 16169070 |
| DSP | DPI | DPII | desmoplakin | - | HPRD,BioGRID | 9261168 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| FABP4 | A-FABP | aP2 | fatty acid binding protein 4, adipocyte | Two-hybrid | BioGRID | 16169070 |
| FAM107A | DRR1 | FLJ30158 | FLJ45473 | TU3A | family with sequence similarity 107, member A | Two-hybrid | BioGRID | 16189514 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | Two-hybrid | BioGRID | 16189514 |
| GOPC | CAL | FIG | GOPC1 | PIST | dJ94G16.2 | golgi associated PDZ and coiled-coil motif containing | Two-hybrid | BioGRID | 16189514 |
| HABP4 | IHABP4 | Ki-1/57 | MGC825 | SERBP1L | hyaluronan binding protein 4 | Two-hybrid | BioGRID | 16169070 |
| HMG20B | BRAF25 | BRAF35 | FLJ26127 | HMGX2 | PP7706 | SMARCE1r | SOXL | pp8857 | high-mobility group 20B | Two-hybrid | BioGRID | 16169070 |
| IVNS1ABP | DKFZp686K06216 | FLARA3 | FLJ10069 | FLJ10411 | FLJ10962 | FLJ35593 | FLJ36593 | HSPC068 | KIAA0850 | ND1 | NS-1 | NS1-BP | NS1BP | influenza virus NS1A binding protein | Two-hybrid | BioGRID | 16169070 |
| KIAA0408 | FLJ43995 | RP3-403A15.2 | KIAA0408 | Two-hybrid | BioGRID | 16189514 |
| KIF15 | FLJ25667 | HKLP2 | KNSL7 | NY-BR-62 | kinesin family member 15 | Two-hybrid | BioGRID | 16169070 |
| KRT20 | CD20 | CK20 | K20 | KRT21 | MGC35423 | keratin 20 | Two-hybrid | BioGRID | 16189514 |
| LOR | MGC111513 | loricrin | Two-hybrid | BioGRID | 16169070 |
| MAFG | MGC13090 | MGC20149 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) | Two-hybrid | BioGRID | 16169070 |
| MAN2A2 | MANA2X | mannosidase, alpha, class 2A, member 2 | Two-hybrid | BioGRID | 16169070 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | menin interacts with vimentin | BIND | 12169273 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | - | HPRD,BioGRID | 12169273 |
| MICAL1 | DKFZp434B1517 | FLJ11937 | FLJ21739 | MICAL | MICAL-1 | NICAL | microtubule associated monoxygenase, calponin and LIM domain containing 1 | - | HPRD,BioGRID | 11827972 |
| MRPL44 | FLJ12701 | FLJ13990 | FLJ37688 | L44MT | MRP-L44 | mitochondrial ribosomal protein L44 | Two-hybrid | BioGRID | 16169070 |
| MYST2 | HBO1 | HBOA | KAT7 | MYST histone acetyltransferase 2 | Two-hybrid | BioGRID | 16169070 |16189514 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | Two-hybrid | BioGRID | 16169070 |
| NIF3L1 | ALS2CR1 | CALS-7 | MDS015 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | Two-hybrid | BioGRID | 16189514 |
| NME2 | MGC111212 | NDPK-B | NDPKB | NM23-H2 | NM23B | puf | non-metastatic cells 2, protein (NM23B) expressed in | - | HPRD,BioGRID | 11082283 |
| NOC4L | MGC3162 | NOC4 | nucleolar complex associated 4 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| NUP85 | FLJ12549 | Nup75 | nucleoporin 85kDa | - | HPRD | 12706117 |
| OSBP2 | ORP-4 | ORP4 | OSBPL1 | OSBPL4 | oxysterol binding protein 2 | - | HPRD,BioGRID | 11802775 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | Two-hybrid | BioGRID | 16189514 |
| PKN1 | DBK | MGC46204 | PAK1 | PKN | PKN-ALPHA | PRK1 | PRKCL1 | protein kinase N1 | - | HPRD,BioGRID | 9175763 |
| PKP1 | B6P | MGC138829 | plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | - | HPRD,BioGRID | 10852826 |
| PLEC1 | EBS1 | EBSO | HD1 | PCN | PLEC1b | PLTN | plectin 1, intermediate filament binding protein 500kDa | - | HPRD,BioGRID | 3027087 |
| PPHLN1 | HSPC206 | HSPC232 | MGC48786 | periphilin 1 | - | HPRD | 15383276 |
| PPL | KIAA0568 | MGC134872 | periplakin | - | HPRD,BioGRID | 12366696 |
| PSMA1 | HC2 | MGC14542 | MGC14575 | MGC14751 | MGC1667 | MGC21459 | MGC22853 | MGC23915 | NU | PROS30 | proteasome (prosome, macropain) subunit, alpha type, 1 | Two-hybrid | BioGRID | 16189514 |
| PSME1 | IFI5111 | MGC8628 | PA28A | PA28alpha | REGalpha | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | Two-hybrid | BioGRID | 16169070 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Two-hybrid | BioGRID | 16169070 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | - | HPRD,BioGRID | 12639940 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | Two-hybrid | BioGRID | 16169070 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| RIBC2 | C22orf11 | RIB43A domain with coiled-coils 2 | Two-hybrid | BioGRID | 16189514 |
| SDCCAG3 | NY-CO-3 | serologically defined colon cancer antigen 3 | Two-hybrid | BioGRID | 16169070 |
| SERBP1 | CGI-55 | CHD3IP | DKFZp564M2423 | FLJ90489 | HABP4L | PAI-RBP1 | PAIRBP1 | SERPINE1 mRNA binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| SH3YL1 | DKFZp586F1318 | FLJ39121 | Ray | SH3 domain containing, Ysc84-like 1 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| SIRT6 | SIR2L6 | sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| SLC25A6 | AAC3 | ANT3 | ANT3Y | MGC17525 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | Two-hybrid | BioGRID | 16169070 |
| SLC27A6 | ACSVL2 | DKFZp779M0564 | FACVL2 | FATP6 | VLCS-H1 | solute carrier family 27 (fatty acid transporter), member 6 | Two-hybrid | BioGRID | 16169070 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | Two-hybrid | BioGRID | 16169070 |
| SUMO2 | HSMT3 | MGC117191 | SMT3B | SMT3H2 | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| SUMO3 | SMT3A | SMT3H1 | SUMO-3 | SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | Two-hybrid | BioGRID | 16169070 |
| TCHP | MGC10854 | trichoplein, keratin filament binding | Two-hybrid | BioGRID | 16189514 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | Two-hybrid | BioGRID | 16169070 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| TRIM28 | FLJ29029 | KAP1 | RNF96 | TF1B | TIF1B | tripartite motif-containing 28 | Two-hybrid | BioGRID | 16169070 |
| TRIM29 | ATDC | FLJ36085 | tripartite motif-containing 29 | - | HPRD | 7644499 |
| TRIOBP | DFNB28 | FLJ39315 | HRIHFB2122 | KIAA1662 | TARA | dJ37E16.4 | TRIO and F-actin binding protein | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| TUBGCP4 | 76P | FLJ14797 | GCP4 | tubulin, gamma complex associated protein 4 | Two-hybrid | BioGRID | 16189514 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | Two-hybrid | BioGRID | 12099690 |
| UPP1 | UDRPASE | UP | UPASE | UPP | uridine phosphorylase 1 | - | HPRD,BioGRID | 11278417 |
| UPP2 | UDRPASE2 | UP2 | UPASE2 | uridine phosphorylase 2 | - | HPRD,BioGRID | 11278417 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 11243787 |11889032 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | - | HPRD | 11375989 |
| XRCC4 | - | X-ray repair complementing defective repair in Chinese hamster cells 4 | Two-hybrid | BioGRID | 16169070 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 10887173 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AURORA B PATHWAY | 39 | 24 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME STRIATED MUSCLE CONTRACTION | 27 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MUSCLE CONTRACTION | 48 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| OHASHI AURKB TARGETS | 10 | 6 | All SZGR 2.0 genes in this pathway |
| HONRADO BREAST CANCER BRCA1 VS BRCA2 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE UP | 15 | 7 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS REVERTED UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER EARLY RECURRENCE | 11 | 10 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 12HR | 35 | 23 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| TING SILENCED BY DICER | 31 | 17 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| MATTHEWS AP1 TARGETS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER HEPATOBLAST | 16 | 12 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| NGO MALIGNANT GLIOMA 1P LOH | 17 | 13 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| MIKHAYLOVA OXIDATIVE STRESS RESPONSE VIA VHL UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS NORMAL EMBRYOS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL DN | 15 | 10 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE ALL DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 81 | 87 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-138 | 32 | 38 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-30-5p | 178 | 184 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.