Gene Page: XRCC1
Summary ?
| GeneID | 7515 |
| Symbol | XRCC1 |
| Synonyms | RCC |
| Description | X-ray repair complementing defective repair in Chinese hamster cells 1 |
| Reference | MIM:194360|HGNC:HGNC:12828|Ensembl:ENSG00000073050|HPRD:01909|Vega:OTTHUMG00000182699 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.802 |
| Sherlock p-value | 0.851 |
| Fetal beta | 0.392 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
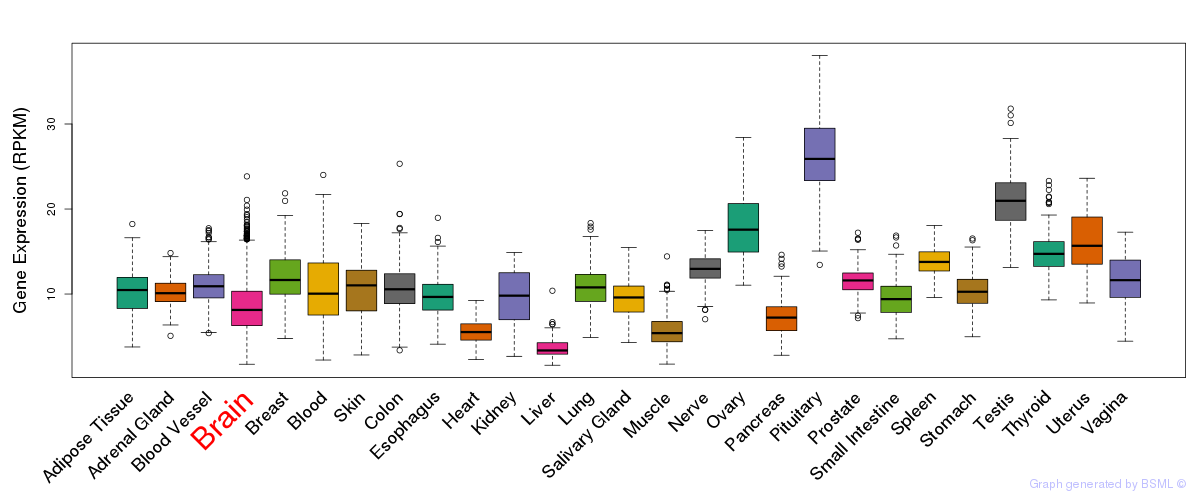
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003684 | damaged DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 14755728 |15044383 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000012 | single strand break repair | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 8978692 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | - | HPRD,BioGRID | 11707423 |
| APTX | AOA | AOA1 | AXA1 | EAOH | EOAHA | FHA-HIT | FLJ20157 | MGC1072 | aprataxin | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15044383 |15555565 |
| APTX | AOA | AOA1 | AXA1 | EAOH | EOAHA | FHA-HIT | FLJ20157 | MGC1072 | aprataxin | APTX interacts with XRCC1. | BIND | 15555565 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 11352725 |
| LIG3 | LIG2 | ligase III, DNA, ATP-dependent | - | HPRD,BioGRID | 8264637 |9136882 |9705932 |11163244 |
| NEIL1 | FLJ22402 | FPG1 | NEI1 | hFPG1 | nei endonuclease VIII-like 1 (E. coli) | XRCC1 interacts with Neil1. | BIND | 15260972 |
| NEIL1 | FLJ22402 | FPG1 | NEI1 | hFPG1 | nei endonuclease VIII-like 1 (E. coli) | - | HPRD,BioGRID | 15260972 |
| OGG1 | HMMH | HOGG1 | MUTM | OGH1 | 8-oxoguanine DNA glycosylase | Reconstituted Complex Two-hybrid | BioGRID | 12933815 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9584196 |15044383 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD | 9584196 |14500814 |
| PARP2 | ADPRT2 | ADPRTL2 | ADPRTL3 | PARP-2 | pADPRT-2 | poly (ADP-ribose) polymerase 2 | Affinity Capture-Western | BioGRID | 11948190 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | Affinity Capture-Western Co-localization FRET Reconstituted Complex | BioGRID | 15107487 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Affinity Capture-MS Affinity Capture-Western Far Western Two-hybrid | BioGRID | 11163244 |17353931 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Phosphorylated XRCC1 interacts with PNK. | BIND | 15066279 |
| POLB | MGC125976 | polymerase (DNA directed), beta | - | HPRD,BioGRID | 8978692 |11467963 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 15044383 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASE EXCISION REPAIR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME BASE EXCISION REPAIR | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 8Q24 DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESISTANCE | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |