Gene Page: XRCC4
Summary ?
| GeneID | 7518 |
| Symbol | XRCC4 |
| Synonyms | SSMED |
| Description | X-ray repair complementing defective repair in Chinese hamster cells 4 |
| Reference | MIM:194363|HGNC:HGNC:12831|Ensembl:ENSG00000152422|HPRD:15951|Vega:OTTHUMG00000131319 |
| Gene type | protein-coding |
| Map location | 5q14.2 |
| Pascal p-value | 0.407 |
| Fetal beta | 0.303 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25243721 | 5 | 82373346 | XRCC4 | -0.019 | 0.74 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
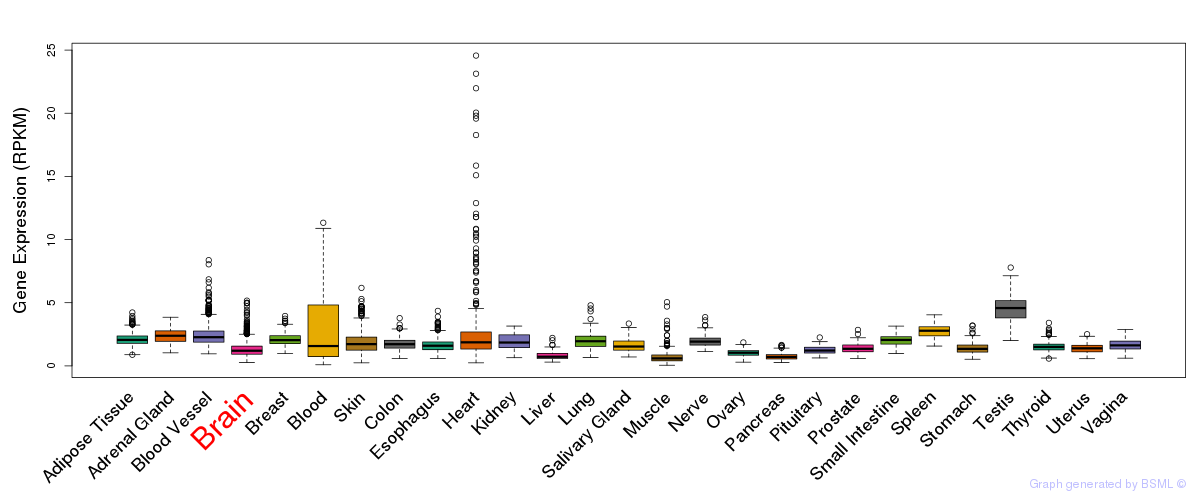
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF583 | 0.94 | 0.95 |
| ZNF564 | 0.94 | 0.91 |
| ZNF180 | 0.93 | 0.93 |
| MED17 | 0.93 | 0.93 |
| C17orf71 | 0.93 | 0.92 |
| ZFP82 | 0.93 | 0.93 |
| ZFP37 | 0.93 | 0.92 |
| ZNF354A | 0.92 | 0.93 |
| TOPORS | 0.92 | 0.92 |
| AC104117.1 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.75 | -0.80 |
| MT-CO2 | -0.74 | -0.86 |
| AF347015.33 | -0.74 | -0.85 |
| AF347015.27 | -0.73 | -0.84 |
| AF347015.31 | -0.73 | -0.84 |
| AIFM3 | -0.73 | -0.79 |
| FXYD1 | -0.72 | -0.85 |
| AF347015.8 | -0.72 | -0.86 |
| MT-CYB | -0.72 | -0.84 |
| S100B | -0.71 | -0.80 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | Two-hybrid | BioGRID | 16189514 |
| AHNAK | AHNAKRS | MGC5395 | AHNAK nucleoprotein | - | HPRD | 15177040 |
| C9orf82 | FLJ13657 | RP11-337A23.1 | chromosome 9 open reading frame 82 | Two-hybrid | BioGRID | 16169070 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| IFFO1 | DKFZp586I2223 | FLJ20703 | HOM-TES-103 | IFFO | MGC117359 | intermediate filament family orphan 1 | Two-hybrid | BioGRID | 16189514 |
| LIG4 | - | ligase IV, DNA, ATP-dependent | DNA Ligase IV (LIG4) interacts with XRCC4. | BIND | 9242410 |
| LIG4 | - | ligase IV, DNA, ATP-dependent | Affinity Capture-Western Co-crystal Structure Two-hybrid | BioGRID | 11702069 |17567543 |
| LIG4 | - | ligase IV, DNA, ATP-dependent | - | HPRD | 10854421 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | Two-hybrid | BioGRID | 18458108 |
| NHEJ1 | FLJ12610 | XLF | nonhomologous end-joining factor 1 | Affinity Capture-Western Two-hybrid | BioGRID | 17567543 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Affinity Capture-MS | BioGRID | 17353931 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD | 9430729 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
| XRCC4 | - | X-ray repair complementing defective repair in Chinese hamster cells 4 | Two-hybrid | BioGRID | 16169070 |17567543 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NON HOMOLOGOUS END JOINING | 14 | 8 | All SZGR 2.0 genes in this pathway |
| PID DNA PK PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME EARLY PHASE OF HIV LIFE CYCLE | 21 | 8 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB UP | 29 | 20 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC REGULATORS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA HP UP | 49 | 26 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN | 50 | 32 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |