Gene Page: ZBTB17
Summary ?
| GeneID | 7709 |
| Symbol | ZBTB17 |
| Synonyms | MIZ-1|ZNF151|ZNF60|pHZ-67 |
| Description | zinc finger and BTB domain containing 17 |
| Reference | MIM:604084|HGNC:HGNC:12936|Ensembl:ENSG00000116809|HPRD:04968|Vega:OTTHUMG00000009377 |
| Gene type | protein-coding |
| Map location | 1p36.13 |
| Pascal p-value | 0.465 |
| Sherlock p-value | 0.813 |
| Fetal beta | 0.397 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07437033 | 1 | 16302232 | ZBTB17 | 5.97E-8 | -0.008 | 1.51E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
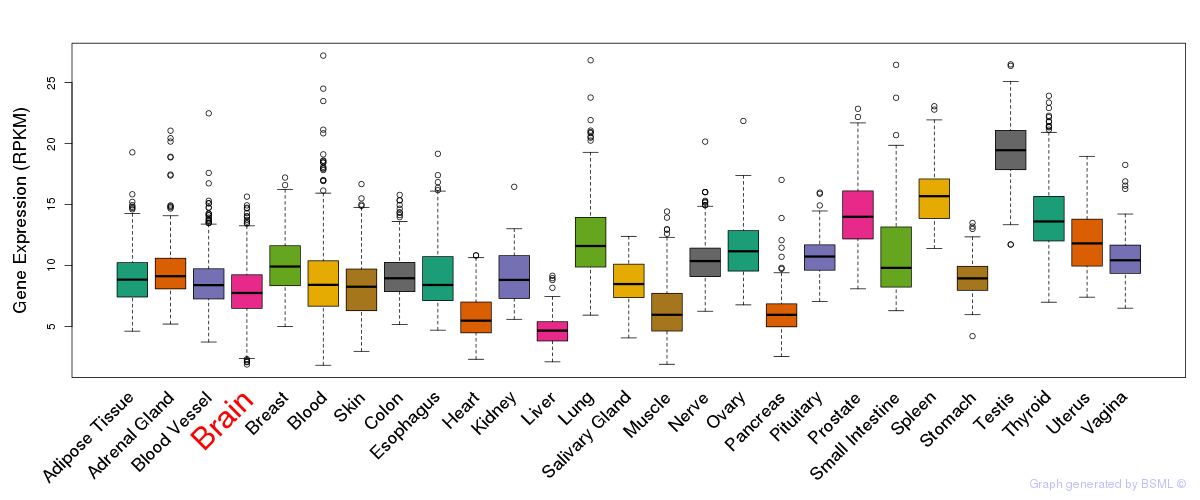
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZCCHC8 | 0.95 | 0.93 |
| RAF1 | 0.95 | 0.95 |
| MARK3 | 0.95 | 0.94 |
| ZNF589 | 0.94 | 0.94 |
| REV1 | 0.94 | 0.94 |
| TTC17 | 0.94 | 0.94 |
| ASXL1 | 0.94 | 0.95 |
| ZNF211 | 0.94 | 0.93 |
| SETDB1 | 0.93 | 0.93 |
| ZNF509 | 0.93 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.92 |
| AF347015.27 | -0.78 | -0.90 |
| MT-CO2 | -0.78 | -0.92 |
| AIFM3 | -0.77 | -0.79 |
| TSC22D4 | -0.76 | -0.82 |
| HLA-F | -0.76 | -0.77 |
| AF347015.33 | -0.76 | -0.88 |
| S100B | -0.76 | -0.84 |
| FXYD1 | -0.76 | -0.88 |
| C5orf53 | -0.76 | -0.76 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Miz1 interacts with p21WAF1 promoter. | BIND | 15856024 |
| CDKN2B | CDK4I | INK4B | MTS2 | P15 | TP15 | p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | - | HPRD | 11283613 |
| DDB2 | DDBB | FLJ34321 | UV-DDB2 | damage-specific DNA binding protein 2, 48kDa | Miz1 interacts with DDB2 promoter. | BIND | 15856024 |
| DIS3 | DKFZp667L1817 | EXOSC11 | FLJ10484 | KIAA1008 | MGC33035 | RP11-342J4.3 | RRP44 | bA555G22.1 | dis3p | DIS3 mitotic control homolog (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | Affinity Capture-Western | BioGRID | 15616584 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | Miz-1 interacts with Dmnt3a. | BIND | 15616584 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 11283613 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD | 12193603 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | - | HPRD,BioGRID | 12244100 |
| HK2 | DKFZp686M1669 | HKII | HXK2 | hexokinase 2 | HKII interacts with Miz-1. This interaction was modeled on a demonstrated interaction between rat HKII and human Miz-1. | BIND | 11068878 |
| HK3 | HKIII | HXK3 | hexokinase 3 (white cell) | - | HPRD,BioGRID | 11068878 |
| HK3 | HKIII | HXK3 | hexokinase 3 (white cell) | HKIII amino-terminal half interacts with Miz-1. This interaction was modeled on a demonstrated interaction between rat HKIII and human Miz-1. | BIND | 11068878 |
| ITGB5 | FLJ26658 | integrin, beta 5 | - | HPRD,BioGRID | 12356872 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | Reconstituted Complex | BioGRID | 11283613 |
| MSX2 | CRS2 | FPP | HOX8 | MSH | PFM | PFM1 | msh homeobox 2 | - | HPRD | 9256341 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 9312026 |
| PMAIP1 | APR | NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | Miz1 interacts with NOXA promoter. | BIND | 15856024 |
| RRM2B | DKFZp686M05248 | MGC102856 | MGC42116 | p53R2 | ribonucleotide reductase M2 B (TP53 inducible) | Miz1 interacts with p53R2 promoter. | BIND | 15856024 |
| TOPBP1 | TOP2BP1 | topoisomerase (DNA) II binding protein 1 | - | HPRD,BioGRID | 12408820 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | 46 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |