Gene Page: CACNB4
Summary ?
| GeneID | 785 |
| Symbol | CACNB4 |
| Synonyms | CAB4|CACNLB4|EA5|EIG9|EJM|EJM4|EJM6 |
| Description | calcium voltage-gated channel auxiliary subunit beta 4 |
| Reference | MIM:601949|HGNC:HGNC:1404|Ensembl:ENSG00000182389|HPRD:09057|Vega:OTTHUMG00000155091 |
| Gene type | protein-coding |
| Map location | 2q23.3 |
| Pascal p-value | 0.058 |
| Sherlock p-value | 0.984 |
| Fetal beta | -1.242 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CANABINOID DOPAMINE EXCITABILITY METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02395 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05105477 | 2 | 152906698 | CACNB4 | 2.2E-5 | 0.606 | 0.017 | DMG:Wockner_2014 |
| cg22159341 | 2 | 152893963 | CACNB4 | 4.786E-4 | 0.7 | 0.046 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | CACNB4 | 785 | 0 | cis | ||
| rs12463708 | chr2 | 149073943 | CACNB4 | 785 | 0.09 | trans | ||
| rs16829545 | chr2 | 151977407 | CACNB4 | 785 | 0.17 | trans | ||
| rs6813178 | chr4 | 71575238 | CACNB4 | 785 | 0.11 | trans | ||
| rs6032295 | chr20 | 44178138 | CACNB4 | 785 | 0.01 | trans |
Section II. Transcriptome annotation
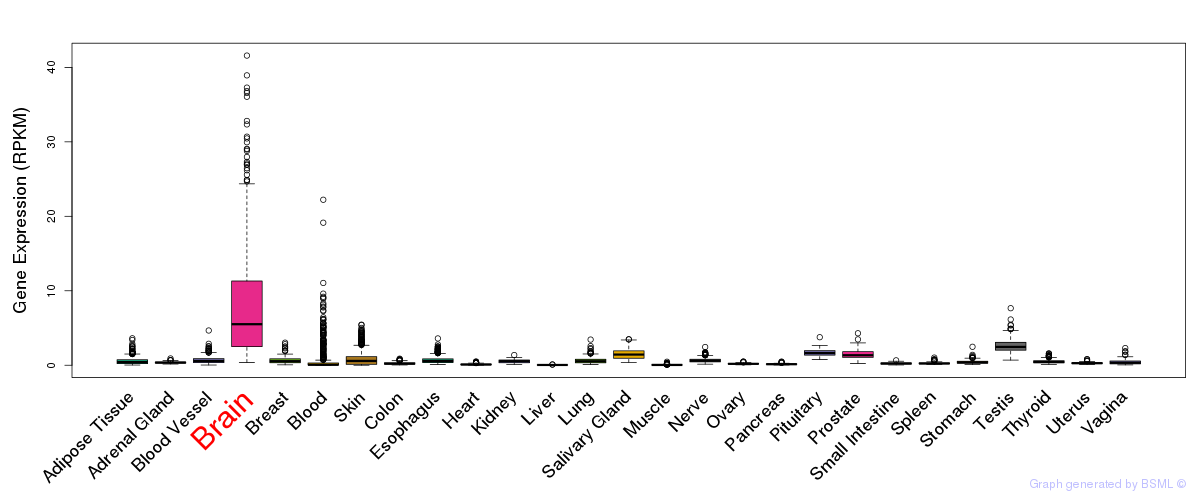
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16525042 | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005245 | voltage-gated calcium channel activity | IDA | 16525042 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0051925 | regulation of calcium ion transport via voltage-gated calcium channel | IDA | 11880487 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8825650 | |
| GO:0009898 | internal side of plasma membrane | TAS | 16385006 | |
| GO:0005891 | voltage-gated calcium channel complex | IDA | 16525042 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM1 INTERACTIONS | 39 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 614 | 620 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-132/212 | 736 | 742 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-151 | 168 | 175 | 1A,m8 | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-182 | 611 | 617 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-183 | 38 | 44 | 1A | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-299-5p | 360 | 366 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-338 | 509 | 515 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-455 | 294 | 301 | 1A,m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-96 | 610 | 617 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.