Gene Page: FZD5
Summary ?
| GeneID | 7855 |
| Symbol | FZD5 |
| Synonyms | C2orf31|HFZ5 |
| Description | frizzled class receptor 5 |
| Reference | MIM:601723|HGNC:HGNC:4043|Ensembl:ENSG00000163251|HPRD:03427|Vega:OTTHUMG00000154790 |
| Gene type | protein-coding |
| Map location | 2q33.3 |
| Pascal p-value | 0.083 |
| Fetal beta | -1.168 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01201797 | 2 | 208635605 | FZD5 | 1.928E-4 | 0.416 | 0.035 | DMG:Wockner_2014 |
| cg05365735 | 2 | 208635637 | FZD5 | 3.122E-4 | 0.449 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | FZD5 | 7855 | 0.05 | trans |
Section II. Transcriptome annotation
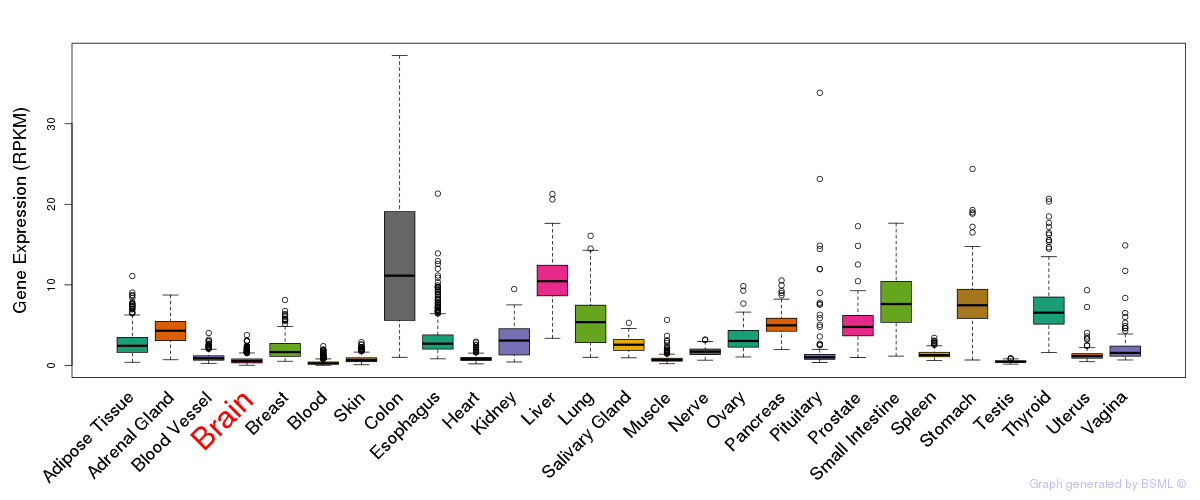
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 9054360 | |
| GO:0004930 | G-protein coupled receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004926 | non-G-protein coupled 7TM receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| GO:0007164 | establishment of tissue polarity | TAS | 8626800 | |
| GO:0048469 | cell maturation | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9054360 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN DEG PATHWAY | 18 | 17 | All SZGR 2.0 genes in this pathway |
| PID WNT SIGNALING PATHWAY | 28 | 23 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS DN | 39 | 20 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 2998 | 3004 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 2997 | 3004 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-140 | 1150 | 1157 | 1A,m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-149 | 1268 | 1274 | 1A | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-155 | 4355 | 4361 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-191 | 77 | 83 | m8 | hsa-miR-191brain | CAACGGAAUCCCAAAAGCAGCU |
| miR-194 | 3738 | 3744 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-219 | 4015 | 4022 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-224 | 3820 | 3826 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-23 | 426 | 433 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24 | 3859 | 3866 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-323 | 426 | 432 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-326 | 3276 | 3282 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG | ||||
| miR-330 | 3387 | 3393 | 1A | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-342 | 3455 | 3461 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-369-3p | 4226 | 4233 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 4227 | 4233 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-375 | 1209 | 1215 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-378* | 3514 | 3521 | 1A,m8 | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-381 | 3296 | 3302 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-384 | 3283 | 3290 | 1A,m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-421 | 3868 | 3875 | 1A,m8 | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-455 | 3872 | 3879 | 1A,m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-99/100 | 3831 | 3837 | m8 | hsa-miR-99abrain | AACCCGUAGAUCCGAUCUUGUG |
| hsa-miR-100brain | AACCCGUAGAUCCGAACUUGUG | ||||
| hsa-miR-99bbrain | CACCCGUAGAACCGACCUUGCG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.