| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP
| 108 | 67 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN
| 431 | 263 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN
| 214 | 133 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN
| 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN
| 129 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN
| 209 | 122 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP
| 40 | 27 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER LARGE VS TINY UP
| 44 | 24 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX
| 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN
| 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP
| 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX4 DN
| 32 | 22 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP
| 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS
| 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215
| 893 | 528 | All SZGR 2.0 genes in this pathway |
| MORI PLASMA CELL UP
| 51 | 29 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP
| 78 | 51 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP
| 197 | 110 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION
| 77 | 62 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK
| 88 | 62 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN
| 121 | 79 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN
| 187 | 122 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN
| 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN
| 337 | 230 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR
| 532 | 309 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP
| 86 | 55 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE
| 64 | 47 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP
| 249 | 170 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP
| 120 | 89 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP
| 93 | 65 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN
| 141 | 92 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP
| 388 | 234 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN
| 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN
| 354 | 216 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP
| 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE
| 89 | 56 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12
| 317 | 177 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN
| 193 | 112 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION
| 368 | 247 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS
| 535 | 325 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2
| 277 | 172 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11
| 103 | 68 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
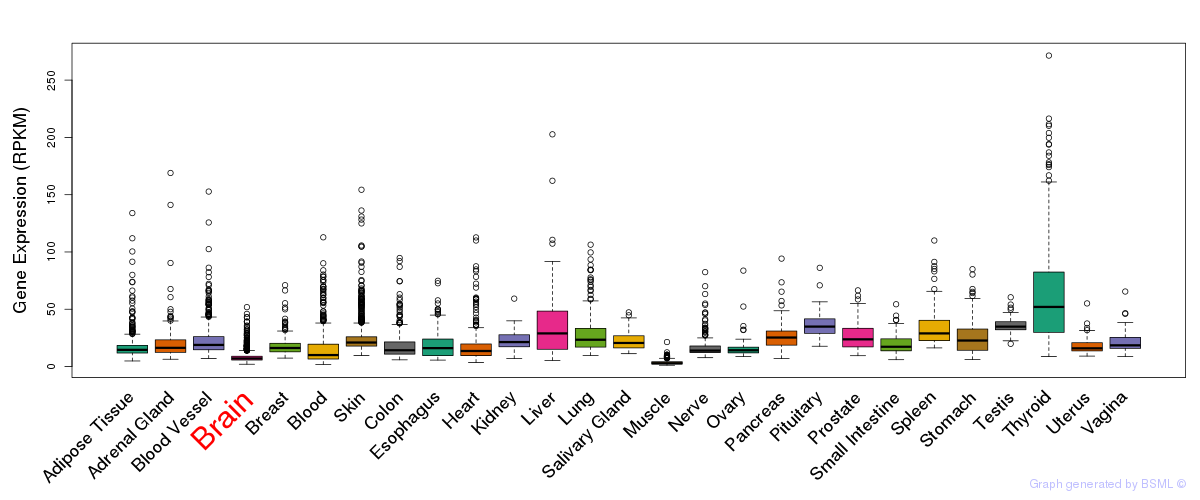
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation