Gene Page: PRRC2A
Summary ?
| GeneID | 7916 |
| Symbol | PRRC2A |
| Synonyms | BAT2|D6S51|D6S51E|G2 |
| Description | proline rich coiled-coil 2A |
| Reference | MIM:142580|HGNC:HGNC:13918|Ensembl:ENSG00000204469|HPRD:00807|Vega:OTTHUMG00000031168 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.319 |
| TADA p-value | 0.006 |
| Fetal beta | 0.061 |
| eGene | Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PRRC2A | chr6 | 31605070..31605071 | TC | T | NM_004638 NM_004638 NM_080686 NM_080686 | . p.2158*>x . p.2158*>x | frameshift read-through frameshift read-through | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
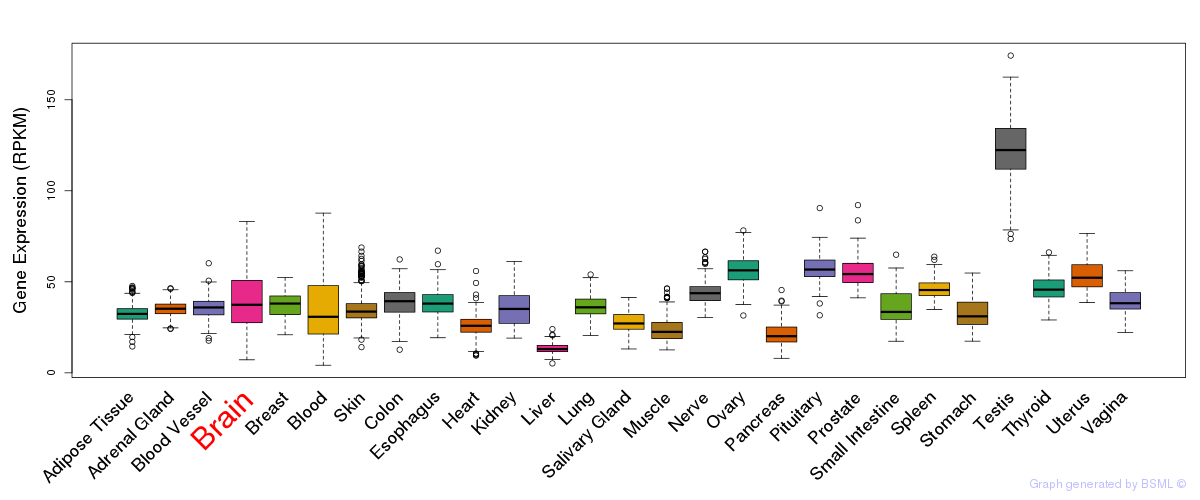
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 14667819 |16713569 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C1QBP | GC1QBP | HABP1 | SF2p32 | gC1Q-R | gC1qR | p32 | complement component 1, q subcomponent binding protein | - | HPRD,BioGRID | 14667819 |
| CCR10 | GPR2 | chemokine (C-C motif) receptor 10 | - | HPRD,BioGRID | 14667819 |
| CPSF1 | CPSF160 | HSU37012 | P/cl.18 | cleavage and polyadenylation specific factor 1, 160kDa | - | HPRD,BioGRID | 14667819 |
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | - | HPRD,BioGRID | 14667819 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | - | HPRD,BioGRID | 14667819 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD,BioGRID | 14667819 |
| GANAB | G2AN | GluII | KIAA0088 | glucosidase, alpha; neutral AB | - | HPRD,BioGRID | 14667819 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 14667819 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | - | HPRD,BioGRID | 14667819 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | - | HPRD,BioGRID | 14667819 |
| IFT88 | D13S1056E | DAF19 | MGC26259 | RP11-172H24.2 | TG737 | TTC10 | hTg737 | intraflagellar transport 88 homolog (Chlamydomonas) | - | HPRD,BioGRID | 14667819 |
| IMMT | DKFZp779P1653 | HMP | MGC111146 | P87 | P87/89 | P89 | PIG4 | PIG52 | inner membrane protein, mitochondrial (mitofilin) | - | HPRD,BioGRID | 14667819 |
| MAD2L2 | MAD2B | REV7 | MAD2 mitotic arrest deficient-like 2 (yeast) | - | HPRD,BioGRID | 14667819 |
| MAN2C1 | DKFZp686E23167 | MAN6A8 | MANA | MANA1 | MGC87979 | mannosidase, alpha, class 2C, member 1 | - | HPRD,BioGRID | 14667819 |
| P4HA2 | - | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II | - | HPRD,BioGRID | 14667819 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | - | HPRD,BioGRID | 14667819 |
| PSMA3 | HC8 | MGC12306 | MGC32631 | PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 | - | HPRD,BioGRID | 14667819 |
| QARS | GLNRS | PRO2195 | glutaminyl-tRNA synthetase | - | HPRD,BioGRID | 14667819 |
| SSR3 | TRAPG | signal sequence receptor, gamma (translocon-associated protein gamma) | - | HPRD,BioGRID | 14667819 |
| UBAP2L | FLJ42300 | KIAA0144 | NICE-4 | ubiquitin associated protein 2-like | - | HPRD,BioGRID | 14667819 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-374 | 42 | 48 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 31 | 38 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-495 | 145 | 152 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.