Gene Page: IRX1
Summary ?
| GeneID | 79192 |
| Symbol | IRX1 |
| Synonyms | IRX-5|IRXA1 |
| Description | iroquois homeobox 1 |
| Reference | MIM:606197|HGNC:HGNC:14358|Ensembl:ENSG00000170549|HPRD:16203|Vega:OTTHUMG00000161632 |
| Gene type | protein-coding |
| Map location | 5p15.3 |
| Pascal p-value | 0.014 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IRX1 | chr5 | 3601122 | C | G | NM_024337 | p.P471A | missense SNV | C | P | Schizophrenia | DNM:McCarthy_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16827839 | chr2 | 150987417 | IRX1 | 79192 | 0.12 | trans | ||
| rs16829545 | chr2 | 151977407 | IRX1 | 79192 | 0.02 | trans | ||
| rs7584986 | chr2 | 184111432 | IRX1 | 79192 | 0.01 | trans | ||
| rs1981925 | chr4 | 34348578 | IRX1 | 79192 | 0.01 | trans | ||
| rs10491487 | chr5 | 80323367 | IRX1 | 79192 | 0.02 | trans | ||
| rs7713523 | chr5 | 97248325 | IRX1 | 79192 | 0.15 | trans | ||
| rs2268637 | chr6 | 39050917 | IRX1 | 79192 | 0.16 | trans | ||
| rs17160028 | chr7 | 31292738 | IRX1 | 79192 | 0.16 | trans | ||
| rs1976192 | chr9 | 117074359 | IRX1 | 79192 | 0.01 | trans | ||
| rs17410931 | chr10 | 98912175 | IRX1 | 79192 | 0.05 | trans | ||
| rs10500981 | chr11 | 24543474 | IRX1 | 79192 | 0.19 | trans | ||
| rs17129612 | chr11 | 96658135 | IRX1 | 79192 | 0.18 | trans | ||
| rs11044062 | chr12 | 8371319 | IRX1 | 79192 | 0.18 | trans | ||
| rs11106191 | chr12 | 78060911 | IRX1 | 79192 | 0.11 | trans | ||
| rs2145444 | chr14 | 52005406 | IRX1 | 79192 | 0.16 | trans | ||
| rs16955618 | chr15 | 29937543 | IRX1 | 79192 | 1.214E-5 | trans | ||
| rs12599961 | chr16 | 8443409 | IRX1 | 79192 | 0.19 | trans | ||
| rs16944856 | chr17 | 11394566 | IRX1 | 79192 | 0.08 | trans | ||
| rs1541352 | chr17 | 13265560 | IRX1 | 79192 | 0.09 | trans | ||
| rs17070739 | chr18 | 60819382 | IRX1 | 79192 | 0.14 | trans | ||
| rs7268640 | chr20 | 47150267 | IRX1 | 79192 | 0.02 | trans |
Section II. Transcriptome annotation
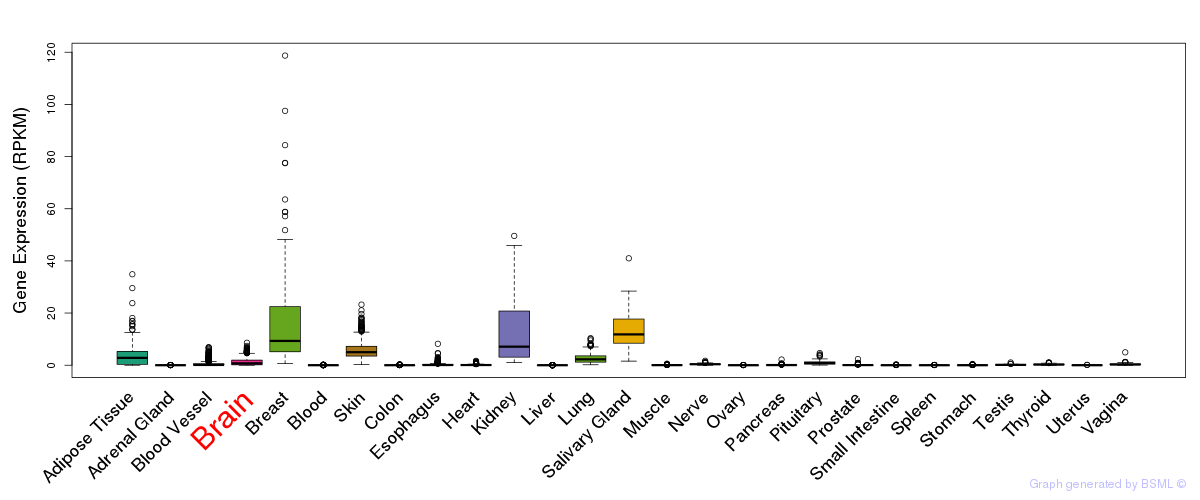
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| GYORFFY MITOXANTRONE RESISTANCE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONIC CARCINOMA VS SEMINOMA DN | 25 | 14 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |