Gene Page: FAT4
Summary ?
| GeneID | 79633 |
| Symbol | FAT4 |
| Synonyms | CDHF14|CDHR11|FAT-J|FATJ|HKLLS2|NBLA00548|VMLDS2 |
| Description | FAT atypical cadherin 4 |
| Reference | MIM:612411|HGNC:HGNC:23109|Ensembl:ENSG00000196159|HPRD:07637|Vega:OTTHUMG00000133100 |
| Gene type | protein-coding |
| Map location | 4q28.1 |
| Pascal p-value | 0.324 |
| Fetal beta | 0.71 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FAT4 | chr4 | 126240143 | A | T | NM_024582 | p.859Q>H | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9288980 | chr3 | 112964991 | FAT4 | 79633 | 0.16 | trans | ||
| rs3995908 | chr3 | 112998529 | FAT4 | 79633 | 0.16 | trans | ||
| rs7652801 | chr3 | 113008605 | FAT4 | 79633 | 0.16 | trans | ||
| rs16889842 | chr4 | 9872012 | FAT4 | 79633 | 0.04 | trans | ||
| rs313946 | chr4 | 113975904 | FAT4 | 79633 | 0.11 | trans | ||
| rs657141 | chr4 | 113981041 | FAT4 | 79633 | 0.13 | trans | ||
| rs2398707 | chr7 | 2113428 | FAT4 | 79633 | 0.19 | trans | ||
| rs3778951 | chr7 | 2125319 | FAT4 | 79633 | 0.19 | trans | ||
| rs3778957 | chr7 | 2127710 | FAT4 | 79633 | 0.19 | trans | ||
| rs3823550 | chr7 | 137346101 | FAT4 | 79633 | 0.19 | trans | ||
| rs7823144 | chr8 | 38028194 | FAT4 | 79633 | 0.18 | trans | ||
| rs9551590 | chr13 | 29589464 | FAT4 | 79633 | 0.16 | trans |
Section II. Transcriptome annotation
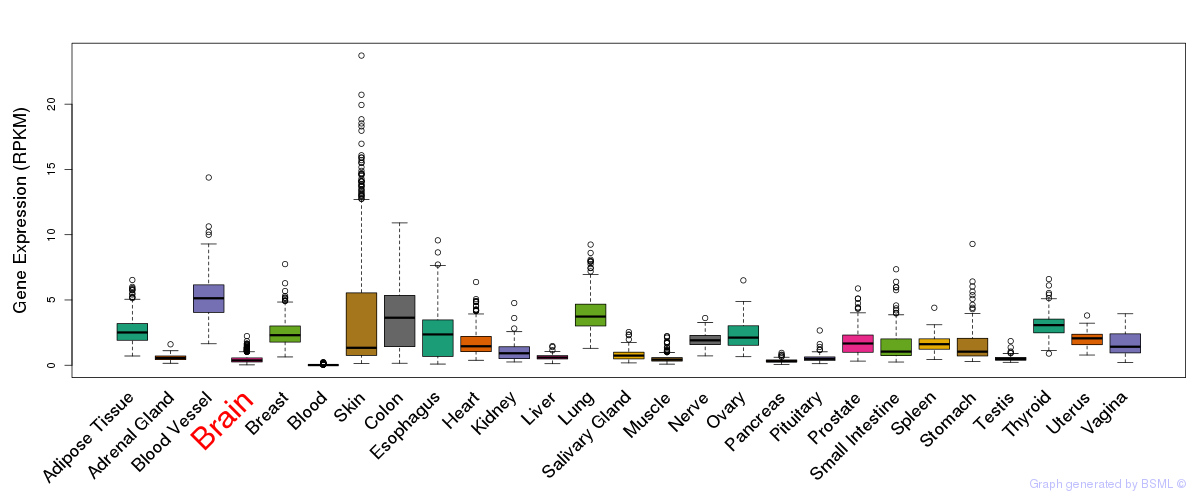
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| CLAUS PGR POSITIVE MENINGIOMA UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL UP | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |