Gene Page: TBL1XR1
Summary ?
| GeneID | 79718 |
| Symbol | TBL1XR1 |
| Synonyms | C21|DC42|IRA1|TBLR1 |
| Description | transducin (beta)-like 1 X-linked receptor 1 |
| Reference | MIM:608628|HGNC:HGNC:29529|Ensembl:ENSG00000177565|HPRD:15476|Vega:OTTHUMG00000157140 |
| Gene type | protein-coding |
| Map location | 3q26.32 |
| Pascal p-value | 0.011 |
| Sherlock p-value | 0.45 |
| Fetal beta | 0.251 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2553600 | chr8 | 133782185 | TBL1XR1 | 79718 | 0.09 | trans | ||
| rs2162141 | chr8 | 133799456 | TBL1XR1 | 79718 | 0.09 | trans | ||
| rs2553588 | chr8 | 133803413 | TBL1XR1 | 79718 | 0.14 | trans | ||
| rs235427 | chr8 | 133815575 | TBL1XR1 | 79718 | 0.01 | trans | ||
| rs235428 | chr8 | 133815725 | TBL1XR1 | 79718 | 0.06 | trans | ||
| rs235436 | chr8 | 133830342 | TBL1XR1 | 79718 | 0.18 | trans | ||
| rs16904746 | chr8 | 133844507 | TBL1XR1 | 79718 | 0.06 | trans | ||
| rs7000440 | chr8 | 133854402 | TBL1XR1 | 79718 | 0.09 | trans | ||
| rs6471100 | 0 | TBL1XR1 | 79718 | 0.07 | trans | |||
| rs7076439 | chr10 | 93286841 | TBL1XR1 | 79718 | 0.12 | trans | ||
| rs11186601 | chr10 | 93287199 | TBL1XR1 | 79718 | 0.1 | trans |
Section II. Transcriptome annotation
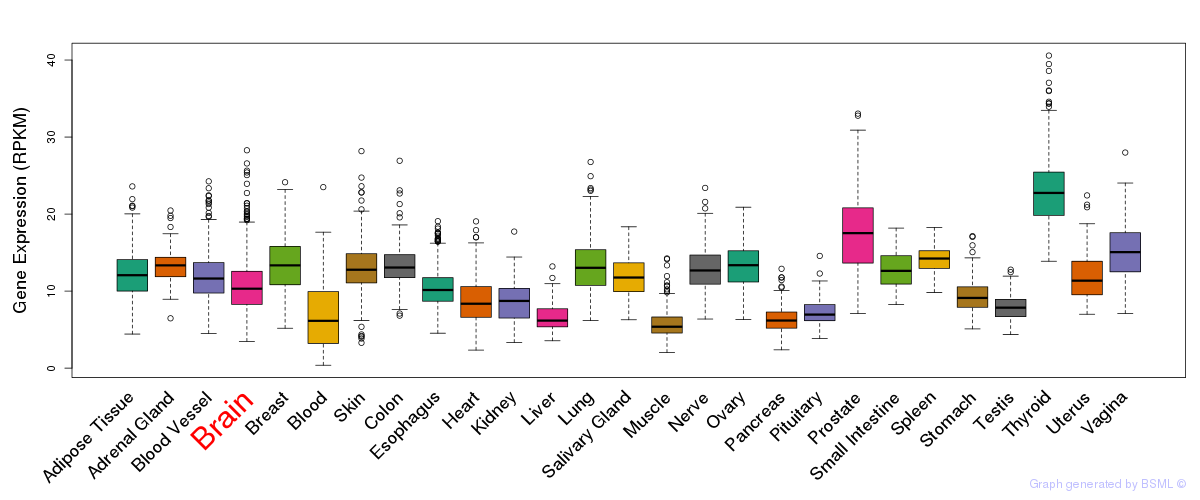
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR2 TARGETS DN | 11 | 6 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |