Gene Page: DCAKD
Summary ?
| GeneID | 79877 |
| Symbol | DCAKD |
| Synonyms | - |
| Description | dephospho-CoA kinase domain containing |
| Reference | HGNC:HGNC:26238|Ensembl:ENSG00000172992|HPRD:08012|Vega:OTTHUMG00000179947 |
| Gene type | protein-coding |
| Map location | 17q21.31 |
| Pascal p-value | 0.039 |
| Sherlock p-value | 0.396 |
| Fetal beta | 0.106 |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
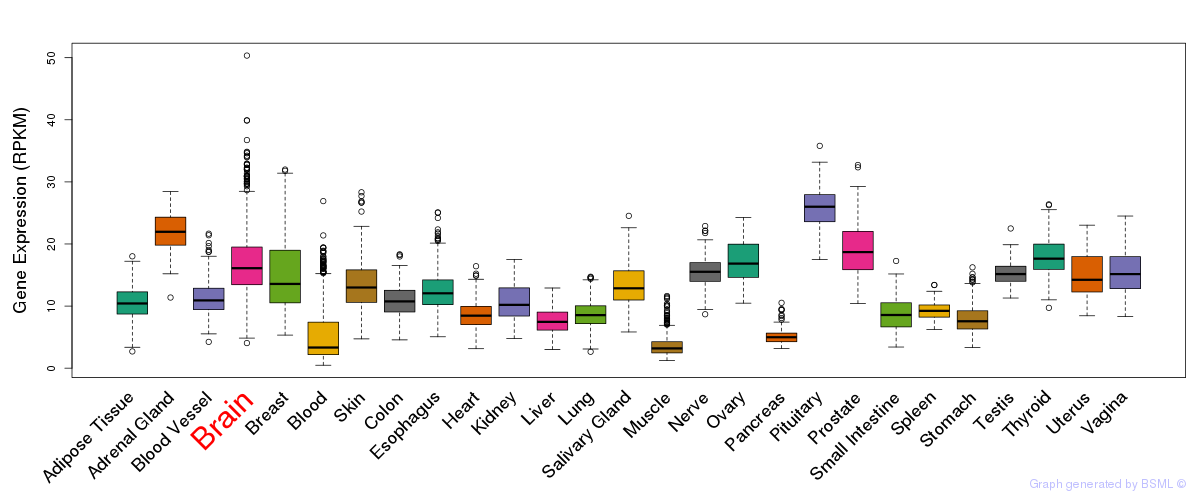
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OLFML3 | 0.57 | 0.36 |
| LUM | 0.57 | 0.43 |
| FCGRT | 0.54 | 0.37 |
| CTSK | 0.53 | 0.35 |
| CTHRC1 | 0.52 | 0.34 |
| MPZL2 | 0.51 | 0.32 |
| C7 | 0.51 | 0.34 |
| ASPN | 0.50 | 0.39 |
| COL3A1 | 0.49 | 0.32 |
| OGN | 0.49 | 0.34 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C19orf68 | -0.26 | -0.25 |
| ZRSR2 | -0.24 | -0.23 |
| AC006435.1 | -0.23 | -0.22 |
| TBC1D2 | -0.23 | -0.20 |
| AC124312.2 | -0.22 | -0.19 |
| MAFK | -0.22 | -0.19 |
| LZTS2 | -0.21 | -0.21 |
| FBXW4 | -0.21 | -0.18 |
| TRIM41 | -0.21 | -0.17 |
| GPATCH4 | -0.21 | -0.18 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA UP | 33 | 28 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |