Gene Page: PIGZ
Summary ?
| GeneID | 80235 |
| Symbol | PIGZ |
| Synonyms | GPI-MT-IV|PIG-Z|SMP3 |
| Description | phosphatidylinositol glycan anchor biosynthesis class Z |
| Reference | MIM:611671|HGNC:HGNC:30596|Ensembl:ENSG00000119227|HPRD:18076|Vega:OTTHUMG00000155522 |
| Gene type | protein-coding |
| Map location | 3q29 |
| Pascal p-value | 0.537 |
| Sherlock p-value | 0.695 |
| Fetal beta | -1.286 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03928961 | 3 | 196693980 | PIGZ | 5.436E-4 | -0.553 | 0.048 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11922888 | 3 | 196675994 | PIGZ | ENSG00000119227.3 | 4.738E-10 | 0 | 19748 | gtex_brain_ba24 |
| rs745738 | 3 | 196676158 | PIGZ | ENSG00000119227.3 | 2.496E-10 | 0 | 19584 | gtex_brain_ba24 |
| rs746037 | 3 | 196676487 | PIGZ | ENSG00000119227.3 | 2.791E-10 | 0 | 19255 | gtex_brain_ba24 |
| rs4347982 | 3 | 196677409 | PIGZ | ENSG00000119227.3 | 7.414E-10 | 0 | 18333 | gtex_brain_ba24 |
| rs9883886 | 3 | 196677481 | PIGZ | ENSG00000119227.3 | 7.414E-10 | 0 | 18261 | gtex_brain_ba24 |
| rs3928150 | 3 | 196678113 | PIGZ | ENSG00000119227.3 | 6.441E-10 | 0 | 17629 | gtex_brain_ba24 |
| rs200475574 | 3 | 196679088 | PIGZ | ENSG00000119227.3 | 1.959E-8 | 0 | 16654 | gtex_brain_ba24 |
| rs4916593 | 3 | 196679090 | PIGZ | ENSG00000119227.3 | 7.425E-9 | 0 | 16652 | gtex_brain_ba24 |
| rs4916594 | 3 | 196679091 | PIGZ | ENSG00000119227.3 | 7.429E-9 | 0 | 16651 | gtex_brain_ba24 |
| rs28498807 | 3 | 196679395 | PIGZ | ENSG00000119227.3 | 3.244E-9 | 0 | 16347 | gtex_brain_ba24 |
| rs6780708 | 3 | 196680730 | PIGZ | ENSG00000119227.3 | 1.039E-9 | 0 | 15012 | gtex_brain_ba24 |
| rs67301616 | 3 | 196681172 | PIGZ | ENSG00000119227.3 | 1.307E-9 | 0 | 14570 | gtex_brain_ba24 |
| rs6808557 | 3 | 196681342 | PIGZ | ENSG00000119227.3 | 1.956E-9 | 0 | 14400 | gtex_brain_ba24 |
| rs11922888 | 3 | 196675994 | PIGZ | ENSG00000119227.3 | 1.055E-10 | 0 | 19748 | gtex_brain_putamen_basal |
| rs745738 | 3 | 196676158 | PIGZ | ENSG00000119227.3 | 5.03E-9 | 0 | 19584 | gtex_brain_putamen_basal |
| rs746037 | 3 | 196676487 | PIGZ | ENSG00000119227.3 | 6.856E-11 | 0 | 19255 | gtex_brain_putamen_basal |
| rs4347982 | 3 | 196677409 | PIGZ | ENSG00000119227.3 | 5.936E-11 | 0 | 18333 | gtex_brain_putamen_basal |
| rs9883886 | 3 | 196677481 | PIGZ | ENSG00000119227.3 | 5.936E-11 | 0 | 18261 | gtex_brain_putamen_basal |
| rs3928150 | 3 | 196678113 | PIGZ | ENSG00000119227.3 | 3.789E-11 | 0 | 17629 | gtex_brain_putamen_basal |
| rs200475574 | 3 | 196679088 | PIGZ | ENSG00000119227.3 | 1.08E-9 | 0 | 16654 | gtex_brain_putamen_basal |
| rs4916593 | 3 | 196679090 | PIGZ | ENSG00000119227.3 | 1.131E-10 | 0 | 16652 | gtex_brain_putamen_basal |
| rs4916594 | 3 | 196679091 | PIGZ | ENSG00000119227.3 | 1.08E-10 | 0 | 16651 | gtex_brain_putamen_basal |
| rs28498807 | 3 | 196679395 | PIGZ | ENSG00000119227.3 | 1.729E-11 | 0 | 16347 | gtex_brain_putamen_basal |
| rs6780708 | 3 | 196680730 | PIGZ | ENSG00000119227.3 | 4.576E-11 | 0 | 15012 | gtex_brain_putamen_basal |
| rs67301616 | 3 | 196681172 | PIGZ | ENSG00000119227.3 | 7.774E-11 | 0 | 14570 | gtex_brain_putamen_basal |
| rs6808557 | 3 | 196681342 | PIGZ | ENSG00000119227.3 | 8.452E-11 | 0 | 14400 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
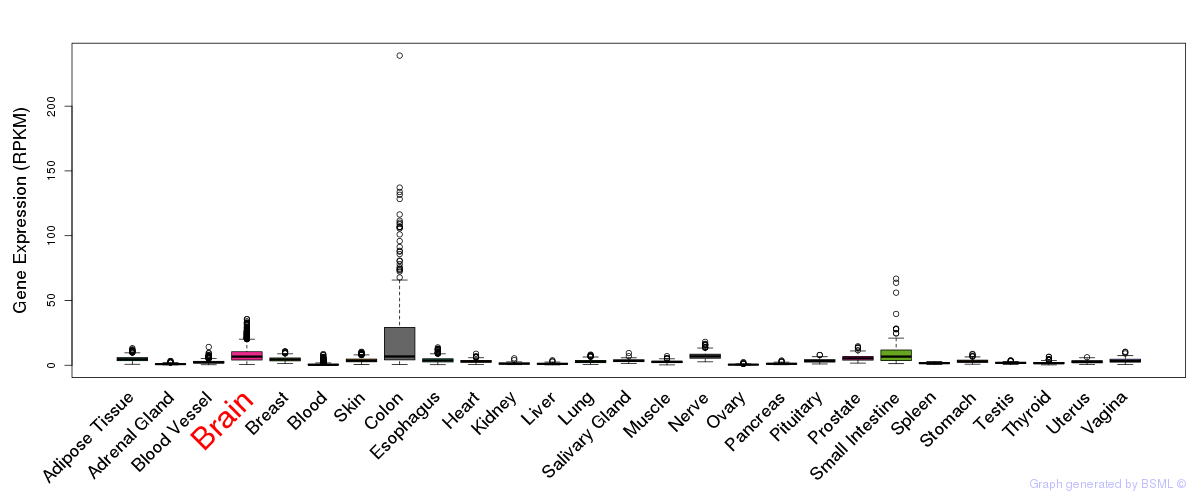
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSYLPHOSPHATIDYLINOSITOL GPI ANCHOR BIOSYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| CHO NR4A1 TARGETS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 2 UP | 54 | 31 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |